laser capture microdissection in pathology
by:Tuowei
2019-09-02
Molecular examination of pathological-altered cells and tissues at DNA, RNA and protein levels has completely altered the study and diagnosis of pathology.
However, the inherent heterogeneity of primary tissues with a mixture of various active cell populations may affect the results and interpretation of molecular studies.
Recently, more and more tissue sections and Microanatomy of cell preparations have been used to separate homogeneous, morphological identified cell populations, thus overcoming obstacles to tissue complexity.
Microanatomy, combined with sensitive analytical techniques, such as PCR, allows accurate in vivo examination of cell populations, such as malignant cells from cancer in situ or hojjkin\'s disease, for traditional molecular research, this is not accessible.
Most microanatomy techniques, however, are time-consuming and require a high degree of manual flexibility, which limits their actual use.
Laser capture microcutting (LCM)
This is a new technology developed by the National Cancer Institute and has made important progress in the speed, ease of use and versatility of microanatomy.
The LCM adheres to the thermoplastic film based on a visually selected cell, the thermoplastic film covers the dehydrated tissue part, and focuses melting by triggering a low energy infrared laser pulse.
The melted film forms a composite with the selected tissue area and can be removed by simply lifting the film.
LCM can be applied to various cell and tissue preparations including paraffin embedded materials.
The use of immune tissue chemical staining allows cells to be selected according to phenotype and functional features.
Depending on the starting material, DNA, high-quality mRNA and protein can be successfully extracted from captured tissue fragments until single cell level.
LCM will combine techniques such as expression library construction, cDNA array hybridization, and differential display to allow the establishment of \"genetic fingerprints\" for specific pathological lesions, especially for malignant tumors \".
In addition to identifying new diagnostic and prognostic markers, this approach helps to establish personalized treatment for tumor molecular features.
This paper outlines the LCM technology, summarizes the current application and new methods, and tries to put forward a prospect for the future development.
In addition, LCM was compared with other newly developed laser microcutting technologies.
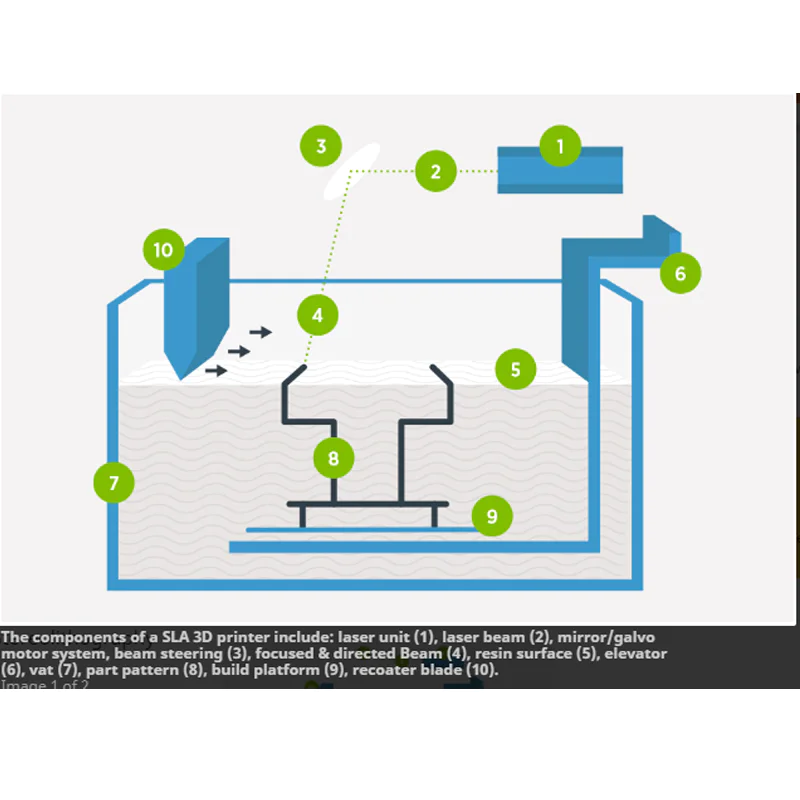
The principle and technical basis of LCMLCM is based on the selective adhesion of visual targeted cells and tissue fragments to thermoplastic films activated by low-energy infrared laser pulses (fig 1).
The system consists of an inverted microscope, solid-state near-infrared laser diode, a laser control unit, a joystick control microscope table with a vacuum chuck for sliding fixing, a CCD camera, and a color display
The LCM microscope is usually connected to a personal computer for additional laser control and image archiving.
The thermoplastic film used to transfer selected batteries is about 6mm in diameter and is mounted on an optical transparent cap that meets standard 0.
5 ml micro centrifuge tubes for further tissue processing.
Download the schematic diagram of figureOpen laser capture microcut in the new tabDownload powerpoint figure 1. (A)
The activation of the laser leads to the melting of the focus of the polymer film. (B)
The lifting of the cap selectively separates cells that adhere to the activated membrane.
The lid is hung on the mechanical transport arm and placed under standard pressure in the desired area of the dehydrated tissue section.
After guiding the visual selection of the required cells by locating the beam, laser activation leads to the focus melting of ethylene vinyl acetate (EVA)
Film, which has the largest absorption near the laser wavelength.
The melted polymer expands to the part, filling the very small hollow space that exists in the tissue.
The polymer breaks down in a few milliseconds and forms a composite with the tissue.
The adhesion of tissue to the activated membrane exceeds the adhesion to the slide and allows selective removal of the required cells.
The laser pulse is usually between 0.
Duration 5 and 5 MS can be repeated multiple times across the surface of the cap, which allows for rapid separation of large numbers of cells.
Harvest selected tissue fragments by simply lifting the lid, and then transfer them to a microcentrifuge tube containing the buffer solution needed to separate the molecules of interest.
At present, the minimum diameter of the laser beam of the LCM microscope is 7.
5 μm with a maximum diameter of 30 μm.
Under standard working conditions, the area of melting of the polymer is exactly the same as the size of the laser point.
Due to the absorption of most of the energy membrane, the maximum temperature reached by the tissue at laser activation is within a range of 90 °c for a few milliseconds, so keep the interested biological large molecules intact.
The low energy of the infrared laser also avoids the potential destruction effect.
Compared with other laser microcutting techniques, the advantages and disadvantages of LCM the most important advantage of LCM is the speed, which combines accuracy and versatility.
Depending on the size of the selected laser point, the structural features of the tissue, and the required micro-anatomical accuracy, thousands of cells can be collected within minutes.
The LCM is several orders of magnitude faster than the micro-anatomy technique based on the micro-manipulator, because it is a \"non-contact\" technique that does not require manual dexterity.
The captured cells and the morphology of the residual tissue are well preserved, allowing the control and recording of tissue representation at all stages of the operation.
Isolated cells are firmly attached to the cap to minimize the risk of tissue loss.
In contrast, in addition to the LMM of laser pressure injection, most other microanatomy techniques require the removal of isolated cells with the help of needles or microcapillary tubes
Unstable steps that require skill and practice.
Since the LCM is very fast and does not destroy adjacent tissues, several tissue components such as normal cells and tumor cells can be sampled sequentially from the same slide.
Due to the simple operation of the LCM microscope, there is actually no time-consuming calibration and adjustment, the learning curve is very short, and the LCM can be easily integrated into the molecular genetic tissue diagnostic program.
LCM can be applied to various cell and tissue preparations.
Even if stained, after removing the coverslip, microanatomy of the archival sections can be successfully performed.
The limitations of the LCM are minimal and basically reflect the difficulty of microanatomy.
While sufficient for most purposes, conventional stained dehydrated tissue slices without coverslips have limited optical resolution and may precisely separate cells from certain complex tissues that lack architectural features, it is almost impossible, such as lymphatic tissue or diffuse invasive cancer.
This problem can be avoided by special stains, especially immune tissue chemistry, which helps to highlight the cell population to be separated or avoided.
The minimum laser spot size of 24 and 25 is 7.
5 μm limits the accuracy of microanatomy of single cells or subcells.
If fragments of adjacent cells are not contaminated, it may be difficult for small cells to separate.
In addition, due to the large tissue contact area of cap, through rare non-
When LCM is performed at the single cell level, specific adhesion cells may be more important.
While LCM was not primarily designed for single cell analysis, several studies have shown that it is feasible to analyze individual or minority cells obtained by LCM from tissue sections and cell preparations.
The latter 26-29 allows LCM to collect pure cell populations faster and more precisely, as cells have been physically separated.
Maitra et al. developed a technique for separating and separating epithelial cell aggregates (EASI)
By spreading pieces of fresh tissue on the slide.
The program separates the epithelial cell complex from the accompanying matrix and allows the LCM to obtain the cells quickly and accurately without the need for tissue slices.
The occasional 28 a problem encountered in slide is the failure to remove the selected cell from the slide.
This may be due to the fact that cells do not adhere to the membrane, usually because the tissue is not completely dehydrated or the laser setting is too low to fully penetrate the melted polymer into the slice.
On the other hand, increasing the adhesion of slices to slides prevents the removal of target cells.
If the frozen section is dried for a long time, this is mostly encountered in the frozen section, while the paraffin section usually does not require special treatment.
Several teams reported detailed protocols to ensure the optimal transfer conditions for a wide range of samples, including those for immune tissue chemical staining.
Another advanced laser micro-cutting technology for commercial use is laser micro-cutting (PALM;
Mikrolaser Marie, Bain Reed, Germany).
Compared to the LCM, with the help of the motorized stage, the tissue is cut out from the surrounding structure by a highly focused UV laser beam.
Tissue can be removed by needle or by laser pressure jet, which allows for free collection of tissue fragments in contact with the lid of the microcentrifuge tube.
15 for many applications, tissue slices are mounted on a thin support film, which is also cut by a laser beam and allows the restoration of large, complete tissue fragments with retained morphology.
Both LCM and LMM are important advances in micro-cutting technology, and future technological developments will improve the performance of these two systems.
Which of these two systems is more suitable for an application depends to a large extent on the requirements of the organization\'s collection.
The high speed, ease of handling, and good control and recording of anatomical tissues make the LCM an ideal tool for quickly collecting large amounts of tissue and bringing together large amounts of single cells.
LMM may be more time consuming and require more skills, but smaller laser beam diameters, maneuvering stages, and large surface areas without tissue contact (
Favorite LCM hat)
It may be more suitable for very precise microanatomy at the single cell and subcell levels.
The application of laser microanatomy although 1976,30 published the first description of laser for tissue microanatomy, these techniques were not widely used until effective analytical methods were introduced for a small number of biological materials.
Microcutting of DNA analysis is an established method that can obtain purified cell populations from fresh and fixed tissues and cell preparations for analyzing genetic changes in DNA levels.
Many different micro-cutting techniques are used to detect loss of impurity (LOH)
In invasive tumors, epithelial lesions of various organs, including the breast, esophagus and prostate, and epithelial lesions in normal tissues adjacent to tumor lesions.
1, 2, 6, 32-34 combined with genome-wide amplification, 17,35 comparative genomic hybridization has been successfully applied to microanatomical tumors and pre-cancerous lesions of the breast, cervix and oral epithelial cells.
36-38loh studies and other DNA-based analyses can easily perform cell and tissue fragments captured by LCM.
16 captured cells were separated from the membrane by enzyme digestion, and if sufficient cells were collected, a standard single-step PCR protocol could be applied.
Due to the small number of tissues used, time-consuming purification steps such as phenol/methane extraction are generally not required.
Analysis of micro-anatomical endocrine tumors plays an important role in the identification of multiple endocrine tumors type 1 (MEN1)
Genes were obtained by location cloning.
39 with newly identified polymorphism markers, single-fold analysis of affected families, and LOH analysis of 200 microanatomical endocrine tumors, the interval was reduced to 300 kb, further analysis of transcripts located in this region in MEN1 kindreds allows the identification of this new tumor-inhibiting gene.
Ovarian has also been used to identify LOH in Ovarian tumors.
The authors found that the proportion of allele loss of chromosome 821 was higher (50%)
Probably because of the uniform cell population obtained by microanatomy.
40 Darling et al analyzed some re-using LCM-
The expression of Gener-type collagen in a patient with a pan-atrophy benign epidermal dissolution bullosa, whose reproductive line is pure, with 2 BP missing in the COL17A1 gene.
They showed that the area was covered by the cutin-forming cells that were re-cut with Type XVII collagen
The expression has additional frame recovery mutations that correct the nonsense-mediated mRNA attenuation, thus creating a mosaic for this gene product.
41 recently, aflatoxin has also been used to demonstrate the intra-tumor sensitivity of the p53 mutation in the mouse lung tumor induced by Huang quyi.
Analysis of malignant Africa by Fend et al
In the same tumor site, there are two hochkin lymphoma with different phenotype and morphology.
Sequencing of the amplified rearranged immunoglobulin genes in two different populations obtained by the LCM of the immune stained slide showed that in all cases there were two unrelated clones, although the PCR analysis of the DNA obtained from the whole slice did not detect a double linearity in any tumor.
25 LCM has also been successfully used for microdissection of Reed-Sternberg (RS)
Cell-like cells from peripheral T-cell lymphoma.
Amplification of immunoglobulin heavy chain genes from collection groups expressing CD20 and abbasstein-microcut RS-like cellsBarr virus (EBV)
Antigen, showing the pattern of multiple-clone or widowed eb-transformed B-cell population unrelated to tumor T-cell cloning.
The latter two studies showed that conventional immune-stained paraffin embedded sections installed on live slides could be used for LCM without interfering with the transfer of captured cells or subsequent PCR analysis.
Because it can quickly sample large amounts of purified cells from heterogeneous tissues, the LCM is also a promising tool for other DNA-based analysis, such as comparative genomic hybridization.
Gene expression analysis tissue or cell-specific gene expression analysis is important in many areas of biological and biomedical research.
Identification of gene expression patterns associated with tumor transformation, inflammation or tissue repair not only has a far-reaching impact on diagnosis and prognosis, but can also prevent medicine by identifying the subclinical stages of the disease, and new treatments for specific gene changes.
43 However, if the total tissue extract is used as a source of mRNA, tissue heterogeneity may make it difficult for the expressed genes to be assigned to a specific cell population.
Confirmation by in situ technique (such as mRNA in situ hybridization or immune tissue chemistry) may not always be possible, which is laborious and time consuming when a large amount of information needs to be checked.
In addition, mRNA in situ hybridization lacks sensitivity for detection of low abundance transcripts.
As a result, many groups have attempted to develop a microcut protocol to produce adequate quality mRNA for subsequent gene expression analysis.
Compared with DNA, mRNA is more sensitive to fixation, is rapidly degraded by the ubiquitous RNase, and requires strict RNase free conditions during sample processing and preparation.
Despite these limitations, several groups recovered high-quality mRNA from microanatomy samples by reverse transcription PCR (RT–PCR)
, Down to the level of single cells.
3, 12, 14, 44 although most authors use fresh tissue, it is reported that, single cells separated by LMM from paraffin embedded archival tissue and fixed metacarn were also successfully embedded in the tissue by paraffin through nested RT-PCR.
However, frozen tissues produce high-quality mRNA and should be used as much as possible to ensure optimal recovery of low abundance information.
MRNA provides several advantages for mRNA analysis.
Its superior speed allows sampling a large number of cells without obvious RNA degradation.
This will help to reduce the illusion caused by a large number of amplification cycles or lack of repeatability due to changes in expression in small cell samples.
In addition, dehydration of tissue slices during LCM may inhibit the activity of tissue rnase, unlike some techniques for microdissection on buffer-covered slices.
Several groups attempted to evaluate the best conditions for the recovery of RNA from tissues receiving LCM.
16,24, 28,46 although the reported protocol varies in some respects, the fixed type, the time in the water medium, the RNase-free conditions in the dyeing and tissue preparation process, the type of organization may also be a related factor.
Usually, the effect of precipitation fixer such as acetone or ethanol is better than that of cross-linking agent such as formaldehyde.
Traditional stains such as sumim/ihong can be carried out in a very short period of time without having a significant impact on RNA recovery.
For many applications, these stains are sufficient for morphological identification and separation of different cell populations for mRNA analysis.
47-49 in order to perform an accurate LCM of tissues showing a lack of structural features, an immune tissue chemical staining protocol has been developed to minimize RNA loss during surgery.
Immune tissue chemistry of frozen slices can be performed in less than 15 minutes, retaining a sufficient number of mRNA to detect cell-specific mRNA from 200 microanatomical cells with 40 amplification cycles
24. Jin et al. used cell rotation and sensitive nested PCR techniques of rat brain tumor cells to amplify brain tumor hormone mRNA from cells captured by individual immune staining.
27 however, compared to conventional staining, immune staining always results in a measurable decrease in RNA recovery, probably due to longer exposure to water Media.
Because the LCM helps to collect precisely determined quantities of purified cells under controlled conditions, its combination with methods such as real-time quantitative RT-PCR will allow for more accurate determination of cell-specific gene expression on microscopic scales.
14,46, 50 in order to better understand the complexity of genetic changes in pathological tissue, it is better to detect multiple different information at the same time than to detect single or minority expressed genes.
Sequence analysis of gene expression or gene chip hybridization can help to establish a characteristic expression map of normal cells and tumor cells.
51-53 gene expression changes related to different steps of tumor occurrence can be revealed by techniques such as differential hybridization or differential display, by comparing related lesions, such as epithelial dysplasia, cancer in situ, and invasive cancer with microanatomy from a single tissue block.
54, 55 Luo et al. demonstrated the feasibility of combining LCM with Gene Chip hybridization, and he showed replicable differences in gene expression between large neurons and small neurons separated from rat dorsal root nodes.
For each experiment, 1000 cells from one population were captured and RNA was amplified with T7 rna pcr to obtain sufficient materials to produce fluorescent probes for microarray hybridization.
26 similar approaches to binding cDNA, cDNA arrays and real-time quantitative PCR were used to show gene expression patterns altered at different stages of breast cancer progression.
As mentioned above, LCM can also be used to obtain expression libraries from purified cell populations of CGAP.
56. There are already several libraries from microanatomical normal, anterior tumor and tumor breast, prostate, ovary, lung and liver tissues on the CGAP website that have a large number of partial sequencing clones (
Recently, studies aimed at identifying prostate-specific genes by analyzing expression sequence tags demonstrate LCM\'s ability to create tissue-specific expression libraries (ESTs).
The highly expressed T-cell receptor γ transcript found in the prostate library generated from the microanatomy tissue was initially thought to be derived from contaminated T cells in the prostate space.
However, subsequent studies have shown that transcripts do come from prostate epithelial cells.
57, 58 similar surprising results, contrary to known tissue expression patterns, can predict the future from this cell-specific genetic analysis method.
In order to produce useful information, the quality of the main tissues used for large-scale expression studies is critical, and organization procurement, freezing time, accuracy of microanatomy, and other parameters need to be strictly controlled.
Single cell analysis although single cell analysis was introduced into pathology and other biomedical research areas before the emergence of laser-based micro-cutting technology, and some amazing progress was made, for example, before implantation, to identify the lineage and cloning of hochkin\'s disease in vitro fertilized egg cells, or to identify genetic diseases, the dedication and manual flexibility required for microanatomy based on micromanipulation hinders the widespread use of this technique.
4, 5, 59, 60 LCM and LMM have been successfully applied to single cell separation.
Mutations in Cancer-causing genes such as RAS, viral DNA sequences, or rearranged immunoglobulin gene sequences were identified from micro-anatomical single cells, gene sequences expressed were successfully amplified from fresh and paraffin-embedded tissues with RT-PCR.
11,12, 14,15, 27,29 Suarez has developed an improved LCM designed for single cell capture
Quian et al replaced the large cover surface with a cylinder covered with EVA polymer film.
21 This reduces the area of contact with the tissue to a band of 40 μm wide, allows for more precise anatomy and greatly reduces non-
Specific adhesion of cells.
By lifting the cylinder with a motor arm and rotating it step by step to provide a new membrane strip, a large number of cells can be collected on the surface, similar to the traditional LCM.
In addition to specific targets, single cells obtained by laser microcutting can also be used as templates for genome-wide amplification, generation of expression libraries, or as probes for expression analysis with a cDNA array.
17,26 single cell analysis raises special questions about microanatomy and subsequent molecular studies.
It is relatively simple to separate single cells from cell preparations with LCM and LMM because cells have been physically separated and are usually complete.
In contrast, the separation of single cells from tissue slices is affected by the tissue properties of the slices and the limits of microanatomy techniques.
Since the optical resolution of the uncovered part is not ideal, although the nucleus is usually identifiable, the cell boundary is blurred in conventional staining.
If the cell compartment (
MRNA, for example)is analysed.
The connected fragments of other cells may greatly alter the expression pattern.
Immune tissue chemical membrane staining can help circumvent this problem, but if the cell profile is irregular, it may not be possible to accurately restore the cell compartment without adjacent cell contamination.
In addition, the slices produce a large number of fragmented cells that lack part of the plasma and even the nucleus.
Therefore, the analysis of the sliced cells can lead to a large loss of genetic information, so many experiments should be carried out.
In addition, the large number of amplification cycles required for single cell analysis increases the risk of artifacts by prioritizing certain templates and contamination.
In addition to these technical problems, single cell results may not represent the entire cell population, such as location in the cell cycle or other random events, due to the analysis of cell-specific events.
Therefore, in many applications, it is advocated to bring together micro-cut single cells to improve the representation of samples and the sensitivity of detection.
Since the LCM is a relatively new technology, improvements and modifications to the general principles of \"aiming and shooting\" can be expected.
When a small number of cells are detected, more precise single cell technology will reduce the possibility of contamination.
21 The combination of an LCM microscope with a computer-controlled programmable stage will make it possible for an automatic LCM of the pre-selected area, further reducing the time required for extensive tissue sampling.
New methods to avoid the fixation of cross-linked fixatives and Tissue embedding, optimization of rapid immune staining protocols, improvement of DNA and RNA extraction techniques will increase the yield of high-quality nucleic acids from small cell samples, and make \"molecular organization\" a reality.
The rich information generated by high-throughput genetic analysis and large-scale sequencing of microanatomical samples using a cDNA microarray will require rigorous verification of other technologies, extracting useful information from diagnosis and prediction will pose a challenge for pathologists and researchers in other fields.
Another dimension of tissue analysis at microscopic scales is the examination of proteins.
Preliminary studies have demonstrated the feasibility of \"Proteomics\" using a two-dimensional gel electrofiner, Western blot, and enzymes on the cell samples obtained by LCM.
16, 61-65 thanks M. Emmert-
Buck read the manuscript critically.
Bianchi AB, navonnnm, Conte CJ.
Detection of loss of impurity in formaldehydeFixed, paraffin-
Tumor specimens were embedded by PCR.
This is J. Pathol 1991; 138:279–84.
The public websites of science Deng G, Lu Y, Zlotnikov G, etc.
There is a lack of impurity in normal tissues next to breast cancer. Science 1996; 274:2057–9.
OpenUrlAbstract/free full text killer hiller T, Snell L, Watson P.
Microscopic anatomical RT-PCR analysis of gene expression in frozen tissues defined by pathology.
Biotechnology 199621:38–44.
Scientific OpenUrlPubMedWeb kankanzler H, Küppers R, Hansmann ML, etc.
Hodgkins and Reed
Sternberg cells in hochkin\'s disease represent (crippled)
B cells in the center. J Exp Med 1996; 184:1495–505.
OpenUrlAbstract/free full Text ü ppers R, Rajewsky K, Zhao M and others.
Hochkin\'s disease: Hodgkins and Reed
Sternberg cells selected from the histological section showed that the cloned immunoglobulin gene was rearranged and appeared to come from B cells at different developmental stages.
Sci Natl Acad Sci usa a 1994; 91:10962–6.
OpenUrlAbstract/free full text radradford DM, Fair K, Thompson AM, etc.
Allele loss of chromosome 17 of breast catheter primary cancer.
Cancer Res 1993; 53:2947–50.
OpenUrlAbstract/free full textwetyml, Maw G, Nadon N, etc.
PCR microanalysis of stained tissue-sliced tumors. Oncogene 1992; 7:2355–61.
Science goes to JJ\'s OpenUrlPubMedWeb, Lamb RF.
Practical histological microanatomy for PCR analysis. J Pathol 1996; 179:121–4.
CA, Cohen, OpenUrlCrossRefPubMedWeb ScienceMoskaluk.
Microcut and PCR amplification of tissue-sliced genomic DNA.
This is J. Pathol 1997; 150:1547–52.
Z, Bertheau, Emmert-Buck MR, et al.
A microcut technique for archival DNA analysis of specific cell populations in lesions of less than 1mm size.
This is J. Pathol 1995; 146:620–5.
OpenUrlPubMedWeb Science I, Becker K-
F, Röhrl MH, etc. Single-
Mutation analysis of tumor cells in stained tissue sections.
Lab investment 199775:801–7.
Mr. johanbernsen of OpenUrlPubMedWeb Science, Homman HBPM, DeVries E, etc.
Identification of multiple mRNA and DNA sequences from laser-isolated small tissue samples
Assistant microscopic anatomy
Lab investment 199878:1267–73.
Scientific OpenUrlPubMedWeb bbhm M, Wieland I, Schütze K, etc.
Micro beam torque: non
Contact laser micro-cutting of film
Natural tissue for installation.
This is J. Pathol 1997; 151:63–7.
L of OpenUrlPubMedWeb Science golf Fink, West W, Ermert L, etc. Real-
Quantitative RT-PCR of time after laser
Selection of auxiliary cellsNat Med 1998; 4:1329–33.
K, Larke of OpenUrlCrossRefPubMedWeb Science deutschü tze.
Identification of expression genes by laser
Mediated single cell operation.
Nat biotechnology 1998; 16:737–42.
Openurlcross Web Science-
Mr. Buck, Bonner RF, Smith PD, etc.
Laser capture microcuttingScience 1996; 274:998–1001.
OpenUrlAbstract/free full Text audio Dietmaier W, Hartmann A, Wallinger S, etc.
Multiple mutation analysis was performed in individual tumor cells with improved genome-wide amplification.
This is J. Pathol 1999; 154:83–95.
RF, Emmert-OpenUrlPubMedWeb Science GmbH Bonner-
Buck M. , Cole K.
Laser capture microcutting: molecular analysis of tissues. Science 1997; 278:1481–3.
OpenUrlFREE full text-
Clower E, vormeyer AO, Bonner RF, etc.
Microanatomy
Gene-based discovery and analysis for cancer progression.
Cancer J Sci Am 1997; 3:259–65.
The Netherlands of OpenUrlPubMedWeb ScienceSimone, Bonner RF, JW Gillespie, etc. Laser-
Capture Microanatomy: Open the microscopic frontier of molecular analysis.
Trends in Genet 1998; 14:272–6.
OpenUrlCrossRefPubMedWeb Science-
Quian CA, Goldstein SR, Pohida T, etc.
Laser capture microscopic cutting of single cells from complex tissues.
Biotechnology 199926:328–35.
RL, Dalca, krauna Road, OpenUrlPubMedWeb Science Limited Strausberg.
New opportunities to reveal the molecular basis of cancer. Nat Genet 1997; 15:S415–16.
Goldstein SR, McQueen PG, Bonner RF.
Thermal Modeling of laser capture micro-cutting.
Applied Optics 199837:7378–91.
F, Emmert-of openurlcrossrefpmedweb Science solar Fend-
Mr. Barker, Chuaqui R, et al. Immuno-
Laser: Laser capture microcutting of immune-stained frozen sections for mRNA analysis.
This is J. Pathol 1999; 154:61–6.
F, Quintanilla, OpenUrlPubMedWeb Science Forum Fend-
Martinez L, Kumar S and others.
Grade B composite
Cell Lymphoma of two cell groups with different immune phenotype is a true double-clone lymphoma.
Molecular analysis of micro-cutting using laser capture.
This is J. Pathol 1999; 154:1857–66.
The science of OpenUrlPubMedWeb luo L, Salunga C, Guo H, et al.
Laser-Gene Expression Map
The adjacent neuron subtypes are captured. Nat Med 1999; 5:117–22.
Scientific openurlcrosspubpubmedweb, Thompson CA, money X, etc.
Analysis of the expression of hormone mRNA in the anterior brain of a single cell with typical features of immune phenotype after laser capture microcutting.
Lab investment 199979:511–12.
OpenUrlPubMedWeb Science, A. Virmani AK, two Virmani AK, and so on.
Enrichment of epithelial cells for molecular research. Nat Med 1999; 5:459–63.
Openurlcrossfood Web Science-
Martinez rose against F, rodrís Moguel L, etc. Peripheral T-
Reed cell lymphomaSternberg-B-like cells
Cell phenotype and genotype associated with Epstein
Bal virus infection
J. Surg Pathol 1999 in the morning; 23:1233–40.
G, Bielser W, Mel-Ruge W, et al.
Laser microscopic cell surgery
Anatomy: A method of preparation. J Microsc 1976; 107:19–24.
OpenUrlPubMedWeb of science
Ruge W, Bielser W, Remy E, etc.
Laser in low temperature freezing micro-cutting technology
Dry tissue slice
Histochem J 1976,8:387–401.
CD of OpenUrlCrossRefPubMedWeb Science audio Vocke, Pozzatti RO, Bostwick DG, etc.
Analysis of 99 cases of prostate microcut cancer showed that the frequency of gene loss in the superior position of the 8p12-21 chromosome was very high.
Cancer Res 1996; 56:2411–16.
OpenUrlAbstract/free full text TextZhuang Z, Murray MJ, Chuaqui R, etc.
In situ microanatomy and invasive breast cancer, the same allele was lost on chromosome 11q13.
Cancer Res 1995; 55:467–71.
OpenUrlAbstract/free full Wenzhuang Z, vormeyer AO, Mark EJ, etc.
Barrett\'s esophagus: cells with a loss of impurity in APC gene sites are pre-clones that invade adenocarcinoma.
Cancer Res 1996; 56:1961–4.
Zhang L, Cui X, Schmidt K, etc.
Genome-wide amplification from individual cells: implications for genetic analysis.
Sci Natl Acad Sci usa a 1992; 89:5847–51.
OpenUrlAbstract/free full Text Aubele M, Zitzelsberger H schenku, etc.
Unique genetic changes of cervical squamous epithelial lesions shown by laser
Assisted Micro-isolation and comparative genomic hybridization. Cancer 1998; 84:375–9.
T, Tenna, Pennanen S and so on of openurlcross refpmedweb sciencukasjarvi.
Comparison of genetic changes in genomic hybridization for detection of breast cancer in the catheter.
This is J. Pathol 1997; 150:1465–71.
Scientific OpenUrlPubMedWeb weber RG, Scheer M, born in IA, etc.
By joint tissue microanatomy, universal DNA amplification, and comparative genomic hybridization, recurrent chromosome imbalances were detected in biopsy materials for pre-oral and malignant lesions.
This is J. Pathol 1998; 153:295–303.
SC, SC guru, Manickam P, of OpenUrlPubMedWeb Science, Chandrasekharappa, etc.
Location cloning of multiple endocrine tumor genestype 1. Science 1997; 276:404–7.
Mr. Brown, Chuaqui R, Vocke CD, etc.
Allele loss on chromosome arm 8 p: Analysis of epithelial ovarian tumors.
Whether Gynecol has 1999; 74:98–102.
TN, Yi C, Bauer JW, etc. of openurlcross refpmedweb Science Limited Darling.
Reply mosaic: partial correction of bacteria
Frame-by-frame mutation in COL17A1-
Recovery mutation
1999 investment in Clin; 103:1371–7.
As a function of OpenUrlPubMedWeb Science saftam, Freet JF, Devero TR, etc.
High-frequency and uneven distribution of p53 mutations in mouse lung tumors induced by Huang Quyi B1.
Cancer Res 1999; 59:3634–40.
OpenUrlAbstract/free full Text paperlieberman R, crovelja, Hawke, etc.
Development of new cancer chemical prophylaxis: the role of drug generation dynamics/efficacy and intermediate endpoint biomarker monitoring. Clin Chem 1998; 44:420–7.
Provide OpenUrlAbstract/free full text to MD, Done SJ, Redston M, etc.
MRNA analysis in microanatomical frozen tissue sections that do not separate RNA.
This is J. Pathol 1998; 153:47–51.
Scientific OpenUrlPubMedWeb shishibitani M, Uneyama C, Miyazaki K, etc.
Methacarn fixation: a new tool for analyzing gene expression in paraffin
Tissue specimens embedded.
Lab investment 200080:199–208.
Scientific OpenUrlPubMedWeb golgoldsworthy SM, Stockton PS, Trempus CS, etc.
Effects of fixation on RNA extraction and amplification of laser capture microtissues.
Moore Carcinog 1999; 25:86–91.
A, Haidian I, grisj, and so on of openurlcrossrefpmedweb Science solar Glasow.
Differential expression of prolactin receptor (PRLR)
In normal and tumor tissues: separation of cell endocrine compartments by laser capture micro-cutting (LCM).
Endocr Res 1998; 24:857–62.
Scientific OpenUrlPubMedWeb A, Haidan A, Hilbers U, etc.
Expression of Ob receptor in normal human adrenaline: different regulation of the functions of the adrenaline cortex and the adrenaline marrow of the hormone.
J. cl in endocrine 1998; 83:4459–66.
Y, Murakami H, Meng Oh, etc. of openurlcross refpmedweb Science.
Analysis of gene expression profile of renal segment in human breast cancer progression.
Kidney Int 2000; 57:321–31.
DC, Teng, robysenk, etc. of openurlcross refpmedweb Science GmbH Sgroi.
In vivo gene expression profile analysis of human breast cancer progression.
Cancer Res 1999; 59:5656–61.
OpenUrlAbstract/free full Text desderisi J, Pan blue, Palm pepper, etc.
Gene expression patterns in human cancer were analyzed using gene chips. Nat Genet 1996; 14:457–60.
Scientific openurlcrosspubmedweb velcuescu VE, Zhang, vogerstein B, etc.
Series analysis of gene expression. Science 1995; 270:484–7.
OpenUrlAbstract/free full Text copy Cossman J, Annunziata cm, in a hurry, etc. Reed-
Sternberg cell genome expression supports B-cell lineage. Blood 1999; 94:411–16.
OpenUrlAbstract/free full Text Luqmani YA, Lymboura mi.
Subtraction hybrid cloning of RNA amplified from different cell populations microcut from tissue sections of low temperature thermostat.
Anal biochemistry 1994; 222:102–9.
P, Paddy AB, OpenUrlCrossRefPubMedWeb Science alimliang.
The difference of messenger RNA was shown by PCR. Science 1992; 257:967–71.
OpenUrlAbstract/free full Text Peterson, Mr Brown, Carlisle AJ, etc.
An improved method for the construction of a directed clone library from micro-cut cells.
Cancer Res 1998; 58:5326–8.
OpenUrlAbstract/free full text vasvasmatzis G, Essand M, Brinkmann U, etc.
Three genes specifically expressed in the human prostate were found by expression sequence tag database analysis.
Sci Natl Acad Sci usa a 1998; 95:300–4.
OpenUrlAbstract/free full text essessand M, Vasmatzis G, Brinkmann U, etc.
High expression of specific T-
Cell Receptor γ transcript in prostate epithelial cells.
Sci Natl Acad Sci usa a 1999; 96:9287–92.
OpenUrlAbstract/free full Text ü ppers R, Zhao Mi, Hansmann ML, etc.
Molecular analysis of single cells selected from histological slices was used to track B cell development in the human reproductive center. EMBO J 1993; 12:4955–67.
Scientific OpenUrlPubMedWeb amd \'Amore F, Stribley JA, Ohno T, etc.
Molecular studies of single cells harvested by micromanipulation from archival tissue sections previously stained with immune tissue chemistry or non-isotope in situ hybridization.
Lab investment 199776:219–24.
OpenUrlPubMed heavy, Deng zhaojiao, Forbes horse, etc.
Laser capture micro-cutting selective acquisition of potential uses of different cell populations for proteomics analysis
Preliminary findings.
1999; 20:689–700.
Openurlcross Web sciencemmert-
Mr. Buck, JW Gillespie, Paweletz CP, etc.
Methods for analysis of tumor proteomics.
Moore Carcinog 2000; 27:158–65.
DK of OpenUrlCrossRefPubMedWeb sciencornstein, englerc, JW Gillespie, etc.
Features of the prostate in cells
Laser capture the specific antigen of the micro-cut benign and malignant prostate epithelial cells.
Clin Cancer Res 2000; 6:353–6.
OpenUrlAbstract/free full TextSimone NL, when Remaley, Charboneau L, etc.
Sensitive immune assay of tissue cell protein obtained by laser capture microcutting.
This is J. Pathol 2000; 156:445–52.
OpenUrlPubMedWeb Science, Y, Lodz caravan, Zhu S, etc. , of Natkanam.
Western blot analysis of 34 expression in histological diversity tumors.
This is J. Pathol 2000; 156:21–7.
However, the inherent heterogeneity of primary tissues with a mixture of various active cell populations may affect the results and interpretation of molecular studies.
Recently, more and more tissue sections and Microanatomy of cell preparations have been used to separate homogeneous, morphological identified cell populations, thus overcoming obstacles to tissue complexity.
Microanatomy, combined with sensitive analytical techniques, such as PCR, allows accurate in vivo examination of cell populations, such as malignant cells from cancer in situ or hojjkin\'s disease, for traditional molecular research, this is not accessible.
Most microanatomy techniques, however, are time-consuming and require a high degree of manual flexibility, which limits their actual use.
Laser capture microcutting (LCM)
This is a new technology developed by the National Cancer Institute and has made important progress in the speed, ease of use and versatility of microanatomy.
The LCM adheres to the thermoplastic film based on a visually selected cell, the thermoplastic film covers the dehydrated tissue part, and focuses melting by triggering a low energy infrared laser pulse.
The melted film forms a composite with the selected tissue area and can be removed by simply lifting the film.
LCM can be applied to various cell and tissue preparations including paraffin embedded materials.
The use of immune tissue chemical staining allows cells to be selected according to phenotype and functional features.
Depending on the starting material, DNA, high-quality mRNA and protein can be successfully extracted from captured tissue fragments until single cell level.
LCM will combine techniques such as expression library construction, cDNA array hybridization, and differential display to allow the establishment of \"genetic fingerprints\" for specific pathological lesions, especially for malignant tumors \".
In addition to identifying new diagnostic and prognostic markers, this approach helps to establish personalized treatment for tumor molecular features.
This paper outlines the LCM technology, summarizes the current application and new methods, and tries to put forward a prospect for the future development.
In addition, LCM was compared with other newly developed laser microcutting technologies.
The principle and technical basis of LCMLCM is based on the selective adhesion of visual targeted cells and tissue fragments to thermoplastic films activated by low-energy infrared laser pulses (fig 1).
The system consists of an inverted microscope, solid-state near-infrared laser diode, a laser control unit, a joystick control microscope table with a vacuum chuck for sliding fixing, a CCD camera, and a color display
The LCM microscope is usually connected to a personal computer for additional laser control and image archiving.
The thermoplastic film used to transfer selected batteries is about 6mm in diameter and is mounted on an optical transparent cap that meets standard 0.
5 ml micro centrifuge tubes for further tissue processing.
Download the schematic diagram of figureOpen laser capture microcut in the new tabDownload powerpoint figure 1. (A)
The activation of the laser leads to the melting of the focus of the polymer film. (B)
The lifting of the cap selectively separates cells that adhere to the activated membrane.
The lid is hung on the mechanical transport arm and placed under standard pressure in the desired area of the dehydrated tissue section.
After guiding the visual selection of the required cells by locating the beam, laser activation leads to the focus melting of ethylene vinyl acetate (EVA)
Film, which has the largest absorption near the laser wavelength.
The melted polymer expands to the part, filling the very small hollow space that exists in the tissue.
The polymer breaks down in a few milliseconds and forms a composite with the tissue.
The adhesion of tissue to the activated membrane exceeds the adhesion to the slide and allows selective removal of the required cells.
The laser pulse is usually between 0.
Duration 5 and 5 MS can be repeated multiple times across the surface of the cap, which allows for rapid separation of large numbers of cells.
Harvest selected tissue fragments by simply lifting the lid, and then transfer them to a microcentrifuge tube containing the buffer solution needed to separate the molecules of interest.
At present, the minimum diameter of the laser beam of the LCM microscope is 7.
5 μm with a maximum diameter of 30 μm.
Under standard working conditions, the area of melting of the polymer is exactly the same as the size of the laser point.
Due to the absorption of most of the energy membrane, the maximum temperature reached by the tissue at laser activation is within a range of 90 °c for a few milliseconds, so keep the interested biological large molecules intact.
The low energy of the infrared laser also avoids the potential destruction effect.
Compared with other laser microcutting techniques, the advantages and disadvantages of LCM the most important advantage of LCM is the speed, which combines accuracy and versatility.
Depending on the size of the selected laser point, the structural features of the tissue, and the required micro-anatomical accuracy, thousands of cells can be collected within minutes.
The LCM is several orders of magnitude faster than the micro-anatomy technique based on the micro-manipulator, because it is a \"non-contact\" technique that does not require manual dexterity.
The captured cells and the morphology of the residual tissue are well preserved, allowing the control and recording of tissue representation at all stages of the operation.
Isolated cells are firmly attached to the cap to minimize the risk of tissue loss.
In contrast, in addition to the LMM of laser pressure injection, most other microanatomy techniques require the removal of isolated cells with the help of needles or microcapillary tubes
Unstable steps that require skill and practice.
Since the LCM is very fast and does not destroy adjacent tissues, several tissue components such as normal cells and tumor cells can be sampled sequentially from the same slide.
Due to the simple operation of the LCM microscope, there is actually no time-consuming calibration and adjustment, the learning curve is very short, and the LCM can be easily integrated into the molecular genetic tissue diagnostic program.
LCM can be applied to various cell and tissue preparations.
Even if stained, after removing the coverslip, microanatomy of the archival sections can be successfully performed.
The limitations of the LCM are minimal and basically reflect the difficulty of microanatomy.
While sufficient for most purposes, conventional stained dehydrated tissue slices without coverslips have limited optical resolution and may precisely separate cells from certain complex tissues that lack architectural features, it is almost impossible, such as lymphatic tissue or diffuse invasive cancer.
This problem can be avoided by special stains, especially immune tissue chemistry, which helps to highlight the cell population to be separated or avoided.
The minimum laser spot size of 24 and 25 is 7.
5 μm limits the accuracy of microanatomy of single cells or subcells.
If fragments of adjacent cells are not contaminated, it may be difficult for small cells to separate.
In addition, due to the large tissue contact area of cap, through rare non-
When LCM is performed at the single cell level, specific adhesion cells may be more important.
While LCM was not primarily designed for single cell analysis, several studies have shown that it is feasible to analyze individual or minority cells obtained by LCM from tissue sections and cell preparations.
The latter 26-29 allows LCM to collect pure cell populations faster and more precisely, as cells have been physically separated.
Maitra et al. developed a technique for separating and separating epithelial cell aggregates (EASI)
By spreading pieces of fresh tissue on the slide.
The program separates the epithelial cell complex from the accompanying matrix and allows the LCM to obtain the cells quickly and accurately without the need for tissue slices.
The occasional 28 a problem encountered in slide is the failure to remove the selected cell from the slide.
This may be due to the fact that cells do not adhere to the membrane, usually because the tissue is not completely dehydrated or the laser setting is too low to fully penetrate the melted polymer into the slice.
On the other hand, increasing the adhesion of slices to slides prevents the removal of target cells.
If the frozen section is dried for a long time, this is mostly encountered in the frozen section, while the paraffin section usually does not require special treatment.
Several teams reported detailed protocols to ensure the optimal transfer conditions for a wide range of samples, including those for immune tissue chemical staining.
Another advanced laser micro-cutting technology for commercial use is laser micro-cutting (PALM;
Mikrolaser Marie, Bain Reed, Germany).
Compared to the LCM, with the help of the motorized stage, the tissue is cut out from the surrounding structure by a highly focused UV laser beam.
Tissue can be removed by needle or by laser pressure jet, which allows for free collection of tissue fragments in contact with the lid of the microcentrifuge tube.
15 for many applications, tissue slices are mounted on a thin support film, which is also cut by a laser beam and allows the restoration of large, complete tissue fragments with retained morphology.
Both LCM and LMM are important advances in micro-cutting technology, and future technological developments will improve the performance of these two systems.
Which of these two systems is more suitable for an application depends to a large extent on the requirements of the organization\'s collection.
The high speed, ease of handling, and good control and recording of anatomical tissues make the LCM an ideal tool for quickly collecting large amounts of tissue and bringing together large amounts of single cells.
LMM may be more time consuming and require more skills, but smaller laser beam diameters, maneuvering stages, and large surface areas without tissue contact (
Favorite LCM hat)
It may be more suitable for very precise microanatomy at the single cell and subcell levels.
The application of laser microanatomy although 1976,30 published the first description of laser for tissue microanatomy, these techniques were not widely used until effective analytical methods were introduced for a small number of biological materials.
Microcutting of DNA analysis is an established method that can obtain purified cell populations from fresh and fixed tissues and cell preparations for analyzing genetic changes in DNA levels.
Many different micro-cutting techniques are used to detect loss of impurity (LOH)
In invasive tumors, epithelial lesions of various organs, including the breast, esophagus and prostate, and epithelial lesions in normal tissues adjacent to tumor lesions.
1, 2, 6, 32-34 combined with genome-wide amplification, 17,35 comparative genomic hybridization has been successfully applied to microanatomical tumors and pre-cancerous lesions of the breast, cervix and oral epithelial cells.
36-38loh studies and other DNA-based analyses can easily perform cell and tissue fragments captured by LCM.
16 captured cells were separated from the membrane by enzyme digestion, and if sufficient cells were collected, a standard single-step PCR protocol could be applied.
Due to the small number of tissues used, time-consuming purification steps such as phenol/methane extraction are generally not required.
Analysis of micro-anatomical endocrine tumors plays an important role in the identification of multiple endocrine tumors type 1 (MEN1)
Genes were obtained by location cloning.
39 with newly identified polymorphism markers, single-fold analysis of affected families, and LOH analysis of 200 microanatomical endocrine tumors, the interval was reduced to 300 kb, further analysis of transcripts located in this region in MEN1 kindreds allows the identification of this new tumor-inhibiting gene.
Ovarian has also been used to identify LOH in Ovarian tumors.
The authors found that the proportion of allele loss of chromosome 821 was higher (50%)
Probably because of the uniform cell population obtained by microanatomy.
40 Darling et al analyzed some re-using LCM-
The expression of Gener-type collagen in a patient with a pan-atrophy benign epidermal dissolution bullosa, whose reproductive line is pure, with 2 BP missing in the COL17A1 gene.
They showed that the area was covered by the cutin-forming cells that were re-cut with Type XVII collagen
The expression has additional frame recovery mutations that correct the nonsense-mediated mRNA attenuation, thus creating a mosaic for this gene product.
41 recently, aflatoxin has also been used to demonstrate the intra-tumor sensitivity of the p53 mutation in the mouse lung tumor induced by Huang quyi.
Analysis of malignant Africa by Fend et al
In the same tumor site, there are two hochkin lymphoma with different phenotype and morphology.
Sequencing of the amplified rearranged immunoglobulin genes in two different populations obtained by the LCM of the immune stained slide showed that in all cases there were two unrelated clones, although the PCR analysis of the DNA obtained from the whole slice did not detect a double linearity in any tumor.
25 LCM has also been successfully used for microdissection of Reed-Sternberg (RS)
Cell-like cells from peripheral T-cell lymphoma.
Amplification of immunoglobulin heavy chain genes from collection groups expressing CD20 and abbasstein-microcut RS-like cellsBarr virus (EBV)
Antigen, showing the pattern of multiple-clone or widowed eb-transformed B-cell population unrelated to tumor T-cell cloning.
The latter two studies showed that conventional immune-stained paraffin embedded sections installed on live slides could be used for LCM without interfering with the transfer of captured cells or subsequent PCR analysis.
Because it can quickly sample large amounts of purified cells from heterogeneous tissues, the LCM is also a promising tool for other DNA-based analysis, such as comparative genomic hybridization.
Gene expression analysis tissue or cell-specific gene expression analysis is important in many areas of biological and biomedical research.
Identification of gene expression patterns associated with tumor transformation, inflammation or tissue repair not only has a far-reaching impact on diagnosis and prognosis, but can also prevent medicine by identifying the subclinical stages of the disease, and new treatments for specific gene changes.
43 However, if the total tissue extract is used as a source of mRNA, tissue heterogeneity may make it difficult for the expressed genes to be assigned to a specific cell population.
Confirmation by in situ technique (such as mRNA in situ hybridization or immune tissue chemistry) may not always be possible, which is laborious and time consuming when a large amount of information needs to be checked.
In addition, mRNA in situ hybridization lacks sensitivity for detection of low abundance transcripts.
As a result, many groups have attempted to develop a microcut protocol to produce adequate quality mRNA for subsequent gene expression analysis.
Compared with DNA, mRNA is more sensitive to fixation, is rapidly degraded by the ubiquitous RNase, and requires strict RNase free conditions during sample processing and preparation.
Despite these limitations, several groups recovered high-quality mRNA from microanatomy samples by reverse transcription PCR (RT–PCR)
, Down to the level of single cells.
3, 12, 14, 44 although most authors use fresh tissue, it is reported that, single cells separated by LMM from paraffin embedded archival tissue and fixed metacarn were also successfully embedded in the tissue by paraffin through nested RT-PCR.
However, frozen tissues produce high-quality mRNA and should be used as much as possible to ensure optimal recovery of low abundance information.
MRNA provides several advantages for mRNA analysis.
Its superior speed allows sampling a large number of cells without obvious RNA degradation.
This will help to reduce the illusion caused by a large number of amplification cycles or lack of repeatability due to changes in expression in small cell samples.
In addition, dehydration of tissue slices during LCM may inhibit the activity of tissue rnase, unlike some techniques for microdissection on buffer-covered slices.
Several groups attempted to evaluate the best conditions for the recovery of RNA from tissues receiving LCM.
16,24, 28,46 although the reported protocol varies in some respects, the fixed type, the time in the water medium, the RNase-free conditions in the dyeing and tissue preparation process, the type of organization may also be a related factor.
Usually, the effect of precipitation fixer such as acetone or ethanol is better than that of cross-linking agent such as formaldehyde.
Traditional stains such as sumim/ihong can be carried out in a very short period of time without having a significant impact on RNA recovery.
For many applications, these stains are sufficient for morphological identification and separation of different cell populations for mRNA analysis.
47-49 in order to perform an accurate LCM of tissues showing a lack of structural features, an immune tissue chemical staining protocol has been developed to minimize RNA loss during surgery.
Immune tissue chemistry of frozen slices can be performed in less than 15 minutes, retaining a sufficient number of mRNA to detect cell-specific mRNA from 200 microanatomical cells with 40 amplification cycles
24. Jin et al. used cell rotation and sensitive nested PCR techniques of rat brain tumor cells to amplify brain tumor hormone mRNA from cells captured by individual immune staining.
27 however, compared to conventional staining, immune staining always results in a measurable decrease in RNA recovery, probably due to longer exposure to water Media.
Because the LCM helps to collect precisely determined quantities of purified cells under controlled conditions, its combination with methods such as real-time quantitative RT-PCR will allow for more accurate determination of cell-specific gene expression on microscopic scales.
14,46, 50 in order to better understand the complexity of genetic changes in pathological tissue, it is better to detect multiple different information at the same time than to detect single or minority expressed genes.
Sequence analysis of gene expression or gene chip hybridization can help to establish a characteristic expression map of normal cells and tumor cells.
51-53 gene expression changes related to different steps of tumor occurrence can be revealed by techniques such as differential hybridization or differential display, by comparing related lesions, such as epithelial dysplasia, cancer in situ, and invasive cancer with microanatomy from a single tissue block.
54, 55 Luo et al. demonstrated the feasibility of combining LCM with Gene Chip hybridization, and he showed replicable differences in gene expression between large neurons and small neurons separated from rat dorsal root nodes.
For each experiment, 1000 cells from one population were captured and RNA was amplified with T7 rna pcr to obtain sufficient materials to produce fluorescent probes for microarray hybridization.
26 similar approaches to binding cDNA, cDNA arrays and real-time quantitative PCR were used to show gene expression patterns altered at different stages of breast cancer progression.
As mentioned above, LCM can also be used to obtain expression libraries from purified cell populations of CGAP.
56. There are already several libraries from microanatomical normal, anterior tumor and tumor breast, prostate, ovary, lung and liver tissues on the CGAP website that have a large number of partial sequencing clones (
Recently, studies aimed at identifying prostate-specific genes by analyzing expression sequence tags demonstrate LCM\'s ability to create tissue-specific expression libraries (ESTs).
The highly expressed T-cell receptor γ transcript found in the prostate library generated from the microanatomy tissue was initially thought to be derived from contaminated T cells in the prostate space.
However, subsequent studies have shown that transcripts do come from prostate epithelial cells.
57, 58 similar surprising results, contrary to known tissue expression patterns, can predict the future from this cell-specific genetic analysis method.
In order to produce useful information, the quality of the main tissues used for large-scale expression studies is critical, and organization procurement, freezing time, accuracy of microanatomy, and other parameters need to be strictly controlled.
Single cell analysis although single cell analysis was introduced into pathology and other biomedical research areas before the emergence of laser-based micro-cutting technology, and some amazing progress was made, for example, before implantation, to identify the lineage and cloning of hochkin\'s disease in vitro fertilized egg cells, or to identify genetic diseases, the dedication and manual flexibility required for microanatomy based on micromanipulation hinders the widespread use of this technique.
4, 5, 59, 60 LCM and LMM have been successfully applied to single cell separation.
Mutations in Cancer-causing genes such as RAS, viral DNA sequences, or rearranged immunoglobulin gene sequences were identified from micro-anatomical single cells, gene sequences expressed were successfully amplified from fresh and paraffin-embedded tissues with RT-PCR.
11,12, 14,15, 27,29 Suarez has developed an improved LCM designed for single cell capture
Quian et al replaced the large cover surface with a cylinder covered with EVA polymer film.
21 This reduces the area of contact with the tissue to a band of 40 μm wide, allows for more precise anatomy and greatly reduces non-
Specific adhesion of cells.
By lifting the cylinder with a motor arm and rotating it step by step to provide a new membrane strip, a large number of cells can be collected on the surface, similar to the traditional LCM.
In addition to specific targets, single cells obtained by laser microcutting can also be used as templates for genome-wide amplification, generation of expression libraries, or as probes for expression analysis with a cDNA array.
17,26 single cell analysis raises special questions about microanatomy and subsequent molecular studies.
It is relatively simple to separate single cells from cell preparations with LCM and LMM because cells have been physically separated and are usually complete.
In contrast, the separation of single cells from tissue slices is affected by the tissue properties of the slices and the limits of microanatomy techniques.
Since the optical resolution of the uncovered part is not ideal, although the nucleus is usually identifiable, the cell boundary is blurred in conventional staining.
If the cell compartment (
MRNA, for example)is analysed.
The connected fragments of other cells may greatly alter the expression pattern.
Immune tissue chemical membrane staining can help circumvent this problem, but if the cell profile is irregular, it may not be possible to accurately restore the cell compartment without adjacent cell contamination.
In addition, the slices produce a large number of fragmented cells that lack part of the plasma and even the nucleus.
Therefore, the analysis of the sliced cells can lead to a large loss of genetic information, so many experiments should be carried out.
In addition, the large number of amplification cycles required for single cell analysis increases the risk of artifacts by prioritizing certain templates and contamination.
In addition to these technical problems, single cell results may not represent the entire cell population, such as location in the cell cycle or other random events, due to the analysis of cell-specific events.
Therefore, in many applications, it is advocated to bring together micro-cut single cells to improve the representation of samples and the sensitivity of detection.
Since the LCM is a relatively new technology, improvements and modifications to the general principles of \"aiming and shooting\" can be expected.
When a small number of cells are detected, more precise single cell technology will reduce the possibility of contamination.
21 The combination of an LCM microscope with a computer-controlled programmable stage will make it possible for an automatic LCM of the pre-selected area, further reducing the time required for extensive tissue sampling.
New methods to avoid the fixation of cross-linked fixatives and Tissue embedding, optimization of rapid immune staining protocols, improvement of DNA and RNA extraction techniques will increase the yield of high-quality nucleic acids from small cell samples, and make \"molecular organization\" a reality.
The rich information generated by high-throughput genetic analysis and large-scale sequencing of microanatomical samples using a cDNA microarray will require rigorous verification of other technologies, extracting useful information from diagnosis and prediction will pose a challenge for pathologists and researchers in other fields.
Another dimension of tissue analysis at microscopic scales is the examination of proteins.
Preliminary studies have demonstrated the feasibility of \"Proteomics\" using a two-dimensional gel electrofiner, Western blot, and enzymes on the cell samples obtained by LCM.
16, 61-65 thanks M. Emmert-
Buck read the manuscript critically.
Bianchi AB, navonnnm, Conte CJ.
Detection of loss of impurity in formaldehydeFixed, paraffin-
Tumor specimens were embedded by PCR.
This is J. Pathol 1991; 138:279–84.
The public websites of science Deng G, Lu Y, Zlotnikov G, etc.
There is a lack of impurity in normal tissues next to breast cancer. Science 1996; 274:2057–9.
OpenUrlAbstract/free full text killer hiller T, Snell L, Watson P.
Microscopic anatomical RT-PCR analysis of gene expression in frozen tissues defined by pathology.
Biotechnology 199621:38–44.
Scientific OpenUrlPubMedWeb kankanzler H, Küppers R, Hansmann ML, etc.
Hodgkins and Reed
Sternberg cells in hochkin\'s disease represent (crippled)
B cells in the center. J Exp Med 1996; 184:1495–505.
OpenUrlAbstract/free full Text ü ppers R, Rajewsky K, Zhao M and others.
Hochkin\'s disease: Hodgkins and Reed
Sternberg cells selected from the histological section showed that the cloned immunoglobulin gene was rearranged and appeared to come from B cells at different developmental stages.
Sci Natl Acad Sci usa a 1994; 91:10962–6.
OpenUrlAbstract/free full text radradford DM, Fair K, Thompson AM, etc.
Allele loss of chromosome 17 of breast catheter primary cancer.
Cancer Res 1993; 53:2947–50.
OpenUrlAbstract/free full textwetyml, Maw G, Nadon N, etc.
PCR microanalysis of stained tissue-sliced tumors. Oncogene 1992; 7:2355–61.
Science goes to JJ\'s OpenUrlPubMedWeb, Lamb RF.
Practical histological microanatomy for PCR analysis. J Pathol 1996; 179:121–4.
CA, Cohen, OpenUrlCrossRefPubMedWeb ScienceMoskaluk.
Microcut and PCR amplification of tissue-sliced genomic DNA.
This is J. Pathol 1997; 150:1547–52.
Z, Bertheau, Emmert-Buck MR, et al.
A microcut technique for archival DNA analysis of specific cell populations in lesions of less than 1mm size.
This is J. Pathol 1995; 146:620–5.
OpenUrlPubMedWeb Science I, Becker K-
F, Röhrl MH, etc. Single-
Mutation analysis of tumor cells in stained tissue sections.
Lab investment 199775:801–7.
Mr. johanbernsen of OpenUrlPubMedWeb Science, Homman HBPM, DeVries E, etc.
Identification of multiple mRNA and DNA sequences from laser-isolated small tissue samples
Assistant microscopic anatomy
Lab investment 199878:1267–73.
Scientific OpenUrlPubMedWeb bbhm M, Wieland I, Schütze K, etc.
Micro beam torque: non
Contact laser micro-cutting of film
Natural tissue for installation.
This is J. Pathol 1997; 151:63–7.
L of OpenUrlPubMedWeb Science golf Fink, West W, Ermert L, etc. Real-
Quantitative RT-PCR of time after laser
Selection of auxiliary cellsNat Med 1998; 4:1329–33.
K, Larke of OpenUrlCrossRefPubMedWeb Science deutschü tze.
Identification of expression genes by laser
Mediated single cell operation.
Nat biotechnology 1998; 16:737–42.
Openurlcross Web Science-
Mr. Buck, Bonner RF, Smith PD, etc.
Laser capture microcuttingScience 1996; 274:998–1001.
OpenUrlAbstract/free full Text audio Dietmaier W, Hartmann A, Wallinger S, etc.
Multiple mutation analysis was performed in individual tumor cells with improved genome-wide amplification.
This is J. Pathol 1999; 154:83–95.
RF, Emmert-OpenUrlPubMedWeb Science GmbH Bonner-
Buck M. , Cole K.
Laser capture microcutting: molecular analysis of tissues. Science 1997; 278:1481–3.
OpenUrlFREE full text-
Clower E, vormeyer AO, Bonner RF, etc.
Microanatomy
Gene-based discovery and analysis for cancer progression.
Cancer J Sci Am 1997; 3:259–65.
The Netherlands of OpenUrlPubMedWeb ScienceSimone, Bonner RF, JW Gillespie, etc. Laser-
Capture Microanatomy: Open the microscopic frontier of molecular analysis.
Trends in Genet 1998; 14:272–6.
OpenUrlCrossRefPubMedWeb Science-
Quian CA, Goldstein SR, Pohida T, etc.
Laser capture microscopic cutting of single cells from complex tissues.
Biotechnology 199926:328–35.
RL, Dalca, krauna Road, OpenUrlPubMedWeb Science Limited Strausberg.
New opportunities to reveal the molecular basis of cancer. Nat Genet 1997; 15:S415–16.
Goldstein SR, McQueen PG, Bonner RF.
Thermal Modeling of laser capture micro-cutting.
Applied Optics 199837:7378–91.
F, Emmert-of openurlcrossrefpmedweb Science solar Fend-
Mr. Barker, Chuaqui R, et al. Immuno-
Laser: Laser capture microcutting of immune-stained frozen sections for mRNA analysis.
This is J. Pathol 1999; 154:61–6.
F, Quintanilla, OpenUrlPubMedWeb Science Forum Fend-
Martinez L, Kumar S and others.
Grade B composite
Cell Lymphoma of two cell groups with different immune phenotype is a true double-clone lymphoma.
Molecular analysis of micro-cutting using laser capture.
This is J. Pathol 1999; 154:1857–66.
The science of OpenUrlPubMedWeb luo L, Salunga C, Guo H, et al.
Laser-Gene Expression Map
The adjacent neuron subtypes are captured. Nat Med 1999; 5:117–22.
Scientific openurlcrosspubpubmedweb, Thompson CA, money X, etc.
Analysis of the expression of hormone mRNA in the anterior brain of a single cell with typical features of immune phenotype after laser capture microcutting.
Lab investment 199979:511–12.
OpenUrlPubMedWeb Science, A. Virmani AK, two Virmani AK, and so on.
Enrichment of epithelial cells for molecular research. Nat Med 1999; 5:459–63.
Openurlcrossfood Web Science-
Martinez rose against F, rodrís Moguel L, etc. Peripheral T-
Reed cell lymphomaSternberg-B-like cells
Cell phenotype and genotype associated with Epstein
Bal virus infection
J. Surg Pathol 1999 in the morning; 23:1233–40.
G, Bielser W, Mel-Ruge W, et al.
Laser microscopic cell surgery
Anatomy: A method of preparation. J Microsc 1976; 107:19–24.
OpenUrlPubMedWeb of science
Ruge W, Bielser W, Remy E, etc.
Laser in low temperature freezing micro-cutting technology
Dry tissue slice
Histochem J 1976,8:387–401.
CD of OpenUrlCrossRefPubMedWeb Science audio Vocke, Pozzatti RO, Bostwick DG, etc.
Analysis of 99 cases of prostate microcut cancer showed that the frequency of gene loss in the superior position of the 8p12-21 chromosome was very high.
Cancer Res 1996; 56:2411–16.
OpenUrlAbstract/free full text TextZhuang Z, Murray MJ, Chuaqui R, etc.
In situ microanatomy and invasive breast cancer, the same allele was lost on chromosome 11q13.
Cancer Res 1995; 55:467–71.
OpenUrlAbstract/free full Wenzhuang Z, vormeyer AO, Mark EJ, etc.
Barrett\'s esophagus: cells with a loss of impurity in APC gene sites are pre-clones that invade adenocarcinoma.
Cancer Res 1996; 56:1961–4.
Zhang L, Cui X, Schmidt K, etc.
Genome-wide amplification from individual cells: implications for genetic analysis.
Sci Natl Acad Sci usa a 1992; 89:5847–51.
OpenUrlAbstract/free full Text Aubele M, Zitzelsberger H schenku, etc.
Unique genetic changes of cervical squamous epithelial lesions shown by laser
Assisted Micro-isolation and comparative genomic hybridization. Cancer 1998; 84:375–9.
T, Tenna, Pennanen S and so on of openurlcross refpmedweb sciencukasjarvi.
Comparison of genetic changes in genomic hybridization for detection of breast cancer in the catheter.
This is J. Pathol 1997; 150:1465–71.
Scientific OpenUrlPubMedWeb weber RG, Scheer M, born in IA, etc.
By joint tissue microanatomy, universal DNA amplification, and comparative genomic hybridization, recurrent chromosome imbalances were detected in biopsy materials for pre-oral and malignant lesions.
This is J. Pathol 1998; 153:295–303.
SC, SC guru, Manickam P, of OpenUrlPubMedWeb Science, Chandrasekharappa, etc.
Location cloning of multiple endocrine tumor genestype 1. Science 1997; 276:404–7.
Mr. Brown, Chuaqui R, Vocke CD, etc.
Allele loss on chromosome arm 8 p: Analysis of epithelial ovarian tumors.
Whether Gynecol has 1999; 74:98–102.
TN, Yi C, Bauer JW, etc. of openurlcross refpmedweb Science Limited Darling.
Reply mosaic: partial correction of bacteria
Frame-by-frame mutation in COL17A1-
Recovery mutation
1999 investment in Clin; 103:1371–7.
As a function of OpenUrlPubMedWeb Science saftam, Freet JF, Devero TR, etc.
High-frequency and uneven distribution of p53 mutations in mouse lung tumors induced by Huang Quyi B1.
Cancer Res 1999; 59:3634–40.
OpenUrlAbstract/free full Text paperlieberman R, crovelja, Hawke, etc.
Development of new cancer chemical prophylaxis: the role of drug generation dynamics/efficacy and intermediate endpoint biomarker monitoring. Clin Chem 1998; 44:420–7.
Provide OpenUrlAbstract/free full text to MD, Done SJ, Redston M, etc.
MRNA analysis in microanatomical frozen tissue sections that do not separate RNA.
This is J. Pathol 1998; 153:47–51.
Scientific OpenUrlPubMedWeb shishibitani M, Uneyama C, Miyazaki K, etc.
Methacarn fixation: a new tool for analyzing gene expression in paraffin
Tissue specimens embedded.
Lab investment 200080:199–208.
Scientific OpenUrlPubMedWeb golgoldsworthy SM, Stockton PS, Trempus CS, etc.
Effects of fixation on RNA extraction and amplification of laser capture microtissues.
Moore Carcinog 1999; 25:86–91.
A, Haidian I, grisj, and so on of openurlcrossrefpmedweb Science solar Glasow.
Differential expression of prolactin receptor (PRLR)
In normal and tumor tissues: separation of cell endocrine compartments by laser capture micro-cutting (LCM).
Endocr Res 1998; 24:857–62.
Scientific OpenUrlPubMedWeb A, Haidan A, Hilbers U, etc.
Expression of Ob receptor in normal human adrenaline: different regulation of the functions of the adrenaline cortex and the adrenaline marrow of the hormone.
J. cl in endocrine 1998; 83:4459–66.
Y, Murakami H, Meng Oh, etc. of openurlcross refpmedweb Science.
Analysis of gene expression profile of renal segment in human breast cancer progression.
Kidney Int 2000; 57:321–31.
DC, Teng, robysenk, etc. of openurlcross refpmedweb Science GmbH Sgroi.
In vivo gene expression profile analysis of human breast cancer progression.
Cancer Res 1999; 59:5656–61.
OpenUrlAbstract/free full Text desderisi J, Pan blue, Palm pepper, etc.
Gene expression patterns in human cancer were analyzed using gene chips. Nat Genet 1996; 14:457–60.
Scientific openurlcrosspubmedweb velcuescu VE, Zhang, vogerstein B, etc.
Series analysis of gene expression. Science 1995; 270:484–7.
OpenUrlAbstract/free full Text copy Cossman J, Annunziata cm, in a hurry, etc. Reed-
Sternberg cell genome expression supports B-cell lineage. Blood 1999; 94:411–16.
OpenUrlAbstract/free full Text Luqmani YA, Lymboura mi.
Subtraction hybrid cloning of RNA amplified from different cell populations microcut from tissue sections of low temperature thermostat.
Anal biochemistry 1994; 222:102–9.
P, Paddy AB, OpenUrlCrossRefPubMedWeb Science alimliang.
The difference of messenger RNA was shown by PCR. Science 1992; 257:967–71.
OpenUrlAbstract/free full Text Peterson, Mr Brown, Carlisle AJ, etc.
An improved method for the construction of a directed clone library from micro-cut cells.
Cancer Res 1998; 58:5326–8.
OpenUrlAbstract/free full text vasvasmatzis G, Essand M, Brinkmann U, etc.
Three genes specifically expressed in the human prostate were found by expression sequence tag database analysis.
Sci Natl Acad Sci usa a 1998; 95:300–4.
OpenUrlAbstract/free full text essessand M, Vasmatzis G, Brinkmann U, etc.
High expression of specific T-
Cell Receptor γ transcript in prostate epithelial cells.
Sci Natl Acad Sci usa a 1999; 96:9287–92.
OpenUrlAbstract/free full Text ü ppers R, Zhao Mi, Hansmann ML, etc.
Molecular analysis of single cells selected from histological slices was used to track B cell development in the human reproductive center. EMBO J 1993; 12:4955–67.
Scientific OpenUrlPubMedWeb amd \'Amore F, Stribley JA, Ohno T, etc.
Molecular studies of single cells harvested by micromanipulation from archival tissue sections previously stained with immune tissue chemistry or non-isotope in situ hybridization.
Lab investment 199776:219–24.
OpenUrlPubMed heavy, Deng zhaojiao, Forbes horse, etc.
Laser capture micro-cutting selective acquisition of potential uses of different cell populations for proteomics analysis
Preliminary findings.
1999; 20:689–700.
Openurlcross Web sciencemmert-
Mr. Buck, JW Gillespie, Paweletz CP, etc.
Methods for analysis of tumor proteomics.
Moore Carcinog 2000; 27:158–65.
DK of OpenUrlCrossRefPubMedWeb sciencornstein, englerc, JW Gillespie, etc.
Features of the prostate in cells
Laser capture the specific antigen of the micro-cut benign and malignant prostate epithelial cells.
Clin Cancer Res 2000; 6:353–6.
OpenUrlAbstract/free full TextSimone NL, when Remaley, Charboneau L, etc.
Sensitive immune assay of tissue cell protein obtained by laser capture microcutting.
This is J. Pathol 2000; 156:445–52.
OpenUrlPubMedWeb Science, Y, Lodz caravan, Zhu S, etc. , of Natkanam.
Western blot analysis of 34 expression in histological diversity tumors.
This is J. Pathol 2000; 156:21–7.
Custom message




 towell@sztuowei.com
towell@sztuowei.com