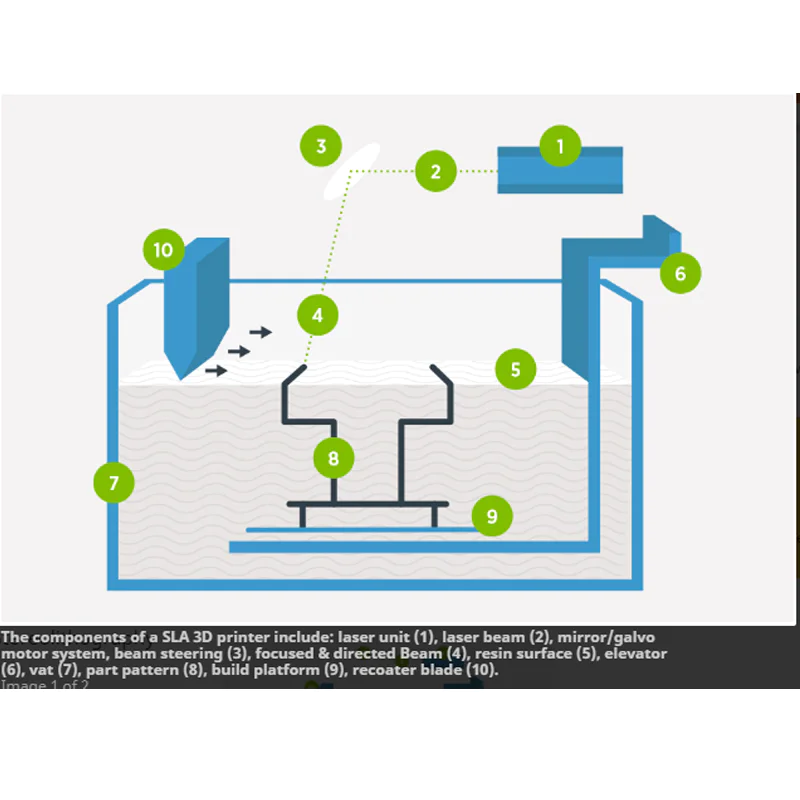
rapid, affordable and portable medium-throughput molecular device for zika virus
by:Tuowei
2019-09-07
Zika virus (ZIKV)
As the cause of fetal microcephaly and Gillan, it has attracted worldwide attention. Barré syndrome.
Existing immunity
Based on rapid detection, it is often impossible to distinguish between Zika virus and associated yellow virus, which are common in affected areas of Central America, South America and the Caribbean.
The Centers for Disease Control and Prevention of the United States and the qualified national health department laboratory can perform reverse transcription PCR (RT-PCR)
ZIKV testing using highly complex instruments with long turnaround times.
Preliminary results for portable and low-
The cost molecular diagnostic system for ZIKV infection was reported.
Less than 15 minutes, this low.
The cost platform can automatically perform high-quality RNA extraction up to 12 ZIKV-
Urine samples were taken at the same time.
Reverse transcription recombinant enzyme PCR amplification reaction can also be carried out (RT-RPA)in ≤15u2009minutes.
Fluorescent signal generated by probebased RT-RPA or RT-
PCR analysis can be monitored using led and smartphone cameras.
Also, RT-RPA and RT-
No cross of PCR analysis
Response to dengue fever and RNA from chikungunya virus. This low-
The cost system lacks complex, sensitive, and costly components,
Limited settings.
It is possible to provide simple samplesto-
Answer the molecular diagnosis and inform the medical staff of the diagnosis in time. ZIKV (
2015 viruses from Mexico)
Harvested in Vero cells 4 days after infection with a concentration of about 2 × 10 plaque-forming units (PFU)/mL.
Then mix the ZIKV suspension with a 3-volume TRIzol reagent (
Science [TFS], Waltham, MA)
The concentration of ZIKV can reach 5 × 10 pfu/mL.
Normal human urine specimens identified (
Directory irann7p)
Research from innovation (Novi, MI).
The modification of the 3D printer is shown in.
Previous studies have shown that the use of bio-Meilie enkerson magnetic extraction reagents and improved consumers can automatically extract DNA and RNA from clinical samples
3D printer (
Printrbot Simple, Printrbot, Inc. , Lincoln, CA).
This 3D printer weighs 12 lbs.
4 lbs. , with a total floor area of 18 \"× 17\" × 13 \"(Lu2009×u2009Wu2009×u2009H).
It can be up to 12 v (6u2009amp)
Computer power supply, which means it can also be powered by a car battery with an inverter.
In the initial design, the continuous binding of MPs with long-sleeved magnets may result in a sub-optimal wash of MPs and/or when RNA (but not DNA)
At a predetermined target.
Therefore, the redesigned magnetic particle processor attachment (MPPA)
Allows for complete MP re-suspension.
In this study, the operation of MPs has made progress for more thorough washing.
Magnetic rods used with tips-
The comb is now in and out of the tip-
Use the extruder motor comb originally designed to move the printed filament to the extruder.
This allows the MPs to be captured and released into different reagents, making the washing step very effective.
MPPA by 3D-
Printed stand attached to luer-
Lock the syringe through two adapters. A disposable 8-tip comb (
Catalogue 97003500, TFS, Waltham, Massachusetts)
Sit safely on the stand.
A set of 8 rod magnet with axial magnetized (
Catalog D2Y0, K & J magnetic materials, Plumsteadville, PA)
The interval of 9mm is fixed in 3D-
A printed plate attached to a metal plate.
The metal plate is made of PLA (PLA)
Filaments with small disk magnets (Catalog R125-
063, amazing magnet, LLC, Anaheim, CA)
Stick to one end.
The vertical movement of the PLA filament, subsequently, the bar magnet controls the extruder stepping motor of the 3D printer through G-codes.
Dynabeads SILANE virus NA kit (TFS)or NucliSENS (bioMérieux)
Magnetic extraction reagents are divided into 96-
Polypropylene deep well plate (Catalog 82007-292, VWR).
When the program is started, the first two lines are deliberately left blank to reconfirm the alignment of the board with MPPA. Wash Buffer 1 (500u2009μL each)
Is added to each hole in line 4 and line 5. Wash Buffer 2 (500u2009μL each)
Added to the well of row 6 and row 7.
Finally, a solution is stored in a 100 µμ l washout buffer or RNA (
Sodium citrate 1 mM, pH 6. 4, TFS)
Added to every well in 12 rows.
In each hole in the third row, the continuously diluted ZIKV sample is 100 μl (
10, 10 and 10)
Mix in human urine with 200 µμ l cracking/binding buffer, 75 µL acetone and 20 µL MP suspension.
Samples were lysed and nucleic acids bound to MPs were performed in the pores at room temperature for 5 minutes. G-
In order to instruct the 3D printer to perform the RNA extraction process, the code was specially written.
Starting from the cracking/bonding hole, MPs are captured when the rod magnet is inserted into the comb and lowered to the hole.
MPPA ships MPs from one well to another, and MPPA is precisely controlled by a 3D printer.
In order to disperse MP particles for thorough washing or washing, the MPPA is shaken vigorously horizontally when the rod magnet is lifted.
Therefore, rna from each sample lysate is bound to MPs, which is washed twice with washing buffer 1 and twice with washing buffer 2, finally, it was eluted in the 100 u2009 μ l eluding buffer or in the RNA storage solution.
The washing steps were performed at 60 °c to improve efficiency.
This heating step is controlled by the heating bed of the 3D printer and the heat of the heating bed is passed to the bottom of the well of the deep well plate using aluminum blocks.
In the standard protocol, the entire 3D printer-
The operation extraction process takes less than 15 min (
5 minutes of cracking and nucleic acid binding, about 6 minutes of washing, 3 minutes of washing).
For comparison, parallel RNA extraction was performed manually using the QIAamp MinElute virus spin kit (
Chagan, Valencia, California)
According to the manufacturer\'s instructions, slightly modified.
In particular, a cell culture of 100 μl-
Series dilution of derivative ZIKV (
10, 10 and 10)
Mix in human urine with 100 µμ l buffer AL and 12.
Qiagen enzyme solution 5 μ l.
After incubation at 56 °c for 15 min, 2.
5 μ l vector RNA solution (
Reserve concentration of 1 μg/μ l in buffer AVE)
Add 95% ml of ethanol to the lysate.
Extract RNA from each dilution in a repeating column and perform standard washing steps as recommended by the manufacturer.
RNA was eluted with 100 μ l buffer AVE at room temperature.
It is worth noting that in both extraction methods, the volume of the input and output samples is deliberately selected to be the same in order to make a fair comparison in efficiency.
RNA templates extracted from our equipment were evaluated in commercial reality
Time PCR detection system (Bio-Rad CFX-
96, Hercules, California)
Platinum One-use superscript III-
Step quantitative RT-PCR system (
TFS, Waltham, MA).
Each 20 μ l reaction consists of a mixture of 10 μ l 2 × Reaction, each 500nm for ZIKV 1086 and ZIKV 1162c primers, FAM-
Labeled hydrolysis probe (ZIKV 1107-FAM)
, And RNA templates of 2 μ l (
Primers and probe sequences see).
The specificity of ZIV primers was previously evaluated by testing dengue, West Nile, Yellow Sea, and chikungunya virus RNA.
The condition is 5 min at 50 °c (cDNA synthesis)
, 95 ° c lasts 30 seconds, 95 ° c lasts 10 seconds, and 60 ° c lasts 20 seconds [
Or known as 300 m/s s/30 m/s s/45 °f × °f (10u2009s/20u2009s)]
Real-time fluorescence detection
Time thermal cycling.
Duration of each business entitytime RT-
PCR runs for about 57 minutes.
When the speed of the commercial Hot Bike is compared to that of the hot water bottle Hot Bike, a shorter protocol is used (TTC)
Developed by the Author: 50 ℃ 30 seconds (cDNA synthesis)
, 95 ° c lasts 10 seconds, 95 ° c lasts 5 seconds, and 60 ° c lasts 10 seconds [30 u2009 s/10 u2009 s/40 u2009 × (5u2009s/10u2009s)].
The duration of the run is 36 minutes and 47 seconds. In most RT-
No PCR reaction
Template controls (NTC)
ZIKV negative controls were added in the same run.
Negative controls are templates in ZIKV negative urine samples or templates containing dengue RNA (DENV-1-4)
Extraction from dengue positive controls (
Heat-inactivated mixture of DENV-1, -2, -3 and -4 standards)
DENV from CDC-1-4 Real Time RT-PCR Assay. For the RT-
In the RPA analysis, we designed the primer and one from the real-time RT-
PCR primers and probes for detecting the prM gene of Zika virus.
RPA primers are 30 ~ according to the recommended length ~ 35 bases.
So rpa zv-F and RPA ZV-
The R primers have 31 and 30 bases respectively, and the size of the amplifier is expected to be 88 ÷ bp ().
The size of Exo stamp exo probe is 52 base lengths ()
, The first 28 bases at the end of probe 5\' overlap with rpa zv-R primer.
The 32 bases at the end of 5\' are thymine, with dT-FAM (
Fluorescent amidite)
, 33 bases at the end of 5\' are Guanyin, replaced by THF (()
, The 34 bases at the end of 5\' are thymine, which is dT-BHQ1 (
Black hole quenching Machine 1).
The design conforms to the standard that THF has at least 30 bases 5\' and THF has at least 15 bases 3.
It also meets the standard of less than 5 bases between fluorescent groups (FAM)
And quenching (BHQ1)
, There is only one secret border of the THF and BHQ1.
Spacers C3 were added at the 3\' end of the probe to prevent the extension. Real-time RT-
According to the manufacturer\'s instructions, the RPA reaction was set using the TwistAmp exo RT kit (
TwistDx, Cambridge, United Kingdom).
Optical clarity 0. 2u2009mL low-
Contour PCR strips with flat covers were used in this trial. For each 12.
5 μ l volume reaction, 420nm for each rpa zv-F and RPA ZV-
The R primer, the 120nm of the TwistAmp exo probe, and the 2 u2009 μ l of the RNA template were mixed and briefly centrifugal, and then the magnesium acetate of 14mm was added to start the reaction.
RT-in this report
RPA is first performed using fluorescence detection in the fluorescence channel (
Excitation 470 nm and detection 520 nm)
In an ESEQuant tubetta (
Konstantz, stockchagan Lake, Germany)
The procedure is maintained at 40 ° c 15 min and the measurement interval is 30 sec.
According to the instructions in the TwistDx protocol, the measurement is suspended at 4 points.
5 min, allow the removal of the reaction tube for a short stirring, then put it back into the tube scanner and immediately resume the fluorescence measurement.
Threshold time (Tt)
The first derivative of the fluorescence signal curve is used to determine the time of the constant temperature amplification reaction.
When the first derivative is greater than or equal to 0.
1. the time point is considered Tt of the analysis.
Instead of using 50 μ l in each RT-
The RPA reaction recommended by the manufacturer is only 12.
5 μ l was used to reduce the cost.
In addition, the smaller reaction volume (e. g. , 5u2009μL)
It is found that no stirring is required in order to achieve the sensitivity claimed by the manufacturer.
Although ESEQuant tubesc is a small portable device that collects fluorescent signals of up to 12 tubes over time, it is also an expensive device (
Over $7,000 per detector).
Thus, the possibility of using a mobile phone as an alternative tool for collecting fluorescent signals was explored. Real-time RT-
Execute RPA on 3D printer-
The processing ZIKV template for the main mixture was incubated using a 3D printer\'s heating bed or extruder as a heat source.
To monitor the green fluorescence of fluorescent dyes, we first tried several blue LED or UV-
The LED flashlight is used as an incentive light source, but the effect is not ideal.
So we built our own LED array (
A row of 8 led separated by 9mm)
Blue LEDs using a separate package (470u2009nm, 5-
Newark 08R2921 diameter mm. com)
As our excitation light source
This special LED model provides high strength (4.
Luminous intensity 1 cd)
Wide Perspective (30°).
The array is powered by a 9 v battery.
In order to capture the green fluorescence emitted by the blue LED lighting reaction tube, the mobile phone (Samsung Note 3)
Plastic orange filter with removal from the gel observation box (
I/O Rodeo, Inc. Pasadena, CA)
The camera placed on the camera lens takes a test tube photo every 30 seconds for $3.
95 Android camera app with lockfocus, locked-
Exposure time-
Delayed photography function (Camera FV-
5 installed on Samsung Note 3 smartphone). The time-
Visual Analysis of failed photos to identify when the signal strength of the tube is greater than ZIKV-
Urine sample control or negative NTC response signal.
We also observed the fluorescence of the tube using a blue LED Gel observation box (
Walkersville, MD).
View and record the launch using the same orange filter and camera.
After the zikv rna template is extracted by the 3D printer, the real-time RT-
PCR using a commercial thermal cycler, via RT-
PCR was performed using TTC.
TTC is a practical solution that can reducecost, simple-to-
Operation and rapid thermal cycling for underserved and developing crowd.
We showed fast PCR and RT-before-
PCR thermal cycling with TTC at a rate of 15 to 30 seconds per cycle.
TTC uses a very simple design based on \"hands-
Transfer the \"reaction tube through a series of water bath liquid to minimize the temperature rise time required for the PCR tube to reach the heat balance.
The automation of PCR reaction is controlled by the action of Arduino micro-controller and servo motor. After the RT-
After the PCR is completed, the same led and smartphone settings as described earlier can be used to view the fluorescent signals of these reaction tubes.
The construction cost per TTC is less than $200.
With TTC, many reactions were carried out in a glass capillary.
The protocol consists of a reversal of 5 sec min, followed by RT inhibition of 10 sec, followed by a degeneration cycle of 40 5 sec and an annealing/extension cycle of 30 sec, abbreviated5u2009s/30u2009s).
Recently, \"I tried a fairly good performance primer to amplify the 30 m/s/10 m/s/40 ÷ × (5u2009s/10u2009s).
It should be noted that when using a glass capillary, the main mixture prepared using the superscript III Platinum 1
Step quantitative RT-PCR system (
Life Technology)
There was no amplification produced, which may have been caused by the adsorption of reverse transcription and PCR on the wall of the glass tube, which effectively reduced the concentration of enzymes that produced the cDNA and the amplification.
So when using a glass tube in TTC, use a step-by-step PrimeScript RT-PCR Kit (
Treasure/Clontech Mountain View, CA)
Compatible with glass tube PCR reaction.
In order to provide the control of the comparison of amplification efficiency, shorter commercial agreements were also carried out.
This 30-inch s/10-inch s/40-hour x-hour (5u2009s/10u2009s)
The commercial thermal cycling reaction takes about 35 minutes to complete.
Expansion of RT-
PCR was evaluated using a 2. 2% pre-casted gel (
Flash gel for longza, volxville, MD)
7 min at 275 V (
4 μ l mixture added to 1 μ l loading dye).
The final gel image was taken with a mobile phone camera.
The typical step sizes used in the gel are 50, 100, 150, 200, 300, 500, 800, and bp.
The image j software developed by NIH quantified the fluorescence intensity of the images taken by the mobile phone camera.
The average gray scale value of the selected fluorescent region is reported.
Differences and spin between Kaijie 3D printer methods
Column method was checked.
Quantitative cycle (Cq)
Using paired t-analyze the values of the two methods
Test with online software (
QuickCalcs, La Jolla, CA for GraphPad). A -
The value is less than 0.
05 is considered to have statistical significance.
The Cq value was also analyzed by the variance equality test of Levene.
As the cause of fetal microcephaly and Gillan, it has attracted worldwide attention. Barré syndrome.
Existing immunity
Based on rapid detection, it is often impossible to distinguish between Zika virus and associated yellow virus, which are common in affected areas of Central America, South America and the Caribbean.
The Centers for Disease Control and Prevention of the United States and the qualified national health department laboratory can perform reverse transcription PCR (RT-PCR)
ZIKV testing using highly complex instruments with long turnaround times.
Preliminary results for portable and low-
The cost molecular diagnostic system for ZIKV infection was reported.
Less than 15 minutes, this low.
The cost platform can automatically perform high-quality RNA extraction up to 12 ZIKV-
Urine samples were taken at the same time.
Reverse transcription recombinant enzyme PCR amplification reaction can also be carried out (RT-RPA)in ≤15u2009minutes.
Fluorescent signal generated by probebased RT-RPA or RT-
PCR analysis can be monitored using led and smartphone cameras.
Also, RT-RPA and RT-
No cross of PCR analysis
Response to dengue fever and RNA from chikungunya virus. This low-
The cost system lacks complex, sensitive, and costly components,
Limited settings.
It is possible to provide simple samplesto-
Answer the molecular diagnosis and inform the medical staff of the diagnosis in time. ZIKV (
2015 viruses from Mexico)
Harvested in Vero cells 4 days after infection with a concentration of about 2 × 10 plaque-forming units (PFU)/mL.
Then mix the ZIKV suspension with a 3-volume TRIzol reagent (
Science [TFS], Waltham, MA)
The concentration of ZIKV can reach 5 × 10 pfu/mL.
Normal human urine specimens identified (
Directory irann7p)
Research from innovation (Novi, MI).
The modification of the 3D printer is shown in.
Previous studies have shown that the use of bio-Meilie enkerson magnetic extraction reagents and improved consumers can automatically extract DNA and RNA from clinical samples
3D printer (
Printrbot Simple, Printrbot, Inc. , Lincoln, CA).
This 3D printer weighs 12 lbs.
4 lbs. , with a total floor area of 18 \"× 17\" × 13 \"(Lu2009×u2009Wu2009×u2009H).
It can be up to 12 v (6u2009amp)
Computer power supply, which means it can also be powered by a car battery with an inverter.
In the initial design, the continuous binding of MPs with long-sleeved magnets may result in a sub-optimal wash of MPs and/or when RNA (but not DNA)
At a predetermined target.
Therefore, the redesigned magnetic particle processor attachment (MPPA)
Allows for complete MP re-suspension.
In this study, the operation of MPs has made progress for more thorough washing.
Magnetic rods used with tips-
The comb is now in and out of the tip-
Use the extruder motor comb originally designed to move the printed filament to the extruder.
This allows the MPs to be captured and released into different reagents, making the washing step very effective.
MPPA by 3D-
Printed stand attached to luer-
Lock the syringe through two adapters. A disposable 8-tip comb (
Catalogue 97003500, TFS, Waltham, Massachusetts)
Sit safely on the stand.
A set of 8 rod magnet with axial magnetized (
Catalog D2Y0, K & J magnetic materials, Plumsteadville, PA)
The interval of 9mm is fixed in 3D-
A printed plate attached to a metal plate.
The metal plate is made of PLA (PLA)
Filaments with small disk magnets (Catalog R125-
063, amazing magnet, LLC, Anaheim, CA)
Stick to one end.
The vertical movement of the PLA filament, subsequently, the bar magnet controls the extruder stepping motor of the 3D printer through G-codes.
Dynabeads SILANE virus NA kit (TFS)or NucliSENS (bioMérieux)
Magnetic extraction reagents are divided into 96-
Polypropylene deep well plate (Catalog 82007-292, VWR).
When the program is started, the first two lines are deliberately left blank to reconfirm the alignment of the board with MPPA. Wash Buffer 1 (500u2009μL each)
Is added to each hole in line 4 and line 5. Wash Buffer 2 (500u2009μL each)
Added to the well of row 6 and row 7.
Finally, a solution is stored in a 100 µμ l washout buffer or RNA (
Sodium citrate 1 mM, pH 6. 4, TFS)
Added to every well in 12 rows.
In each hole in the third row, the continuously diluted ZIKV sample is 100 μl (
10, 10 and 10)
Mix in human urine with 200 µμ l cracking/binding buffer, 75 µL acetone and 20 µL MP suspension.
Samples were lysed and nucleic acids bound to MPs were performed in the pores at room temperature for 5 minutes. G-
In order to instruct the 3D printer to perform the RNA extraction process, the code was specially written.
Starting from the cracking/bonding hole, MPs are captured when the rod magnet is inserted into the comb and lowered to the hole.
MPPA ships MPs from one well to another, and MPPA is precisely controlled by a 3D printer.
In order to disperse MP particles for thorough washing or washing, the MPPA is shaken vigorously horizontally when the rod magnet is lifted.
Therefore, rna from each sample lysate is bound to MPs, which is washed twice with washing buffer 1 and twice with washing buffer 2, finally, it was eluted in the 100 u2009 μ l eluding buffer or in the RNA storage solution.
The washing steps were performed at 60 °c to improve efficiency.
This heating step is controlled by the heating bed of the 3D printer and the heat of the heating bed is passed to the bottom of the well of the deep well plate using aluminum blocks.
In the standard protocol, the entire 3D printer-
The operation extraction process takes less than 15 min (
5 minutes of cracking and nucleic acid binding, about 6 minutes of washing, 3 minutes of washing).
For comparison, parallel RNA extraction was performed manually using the QIAamp MinElute virus spin kit (
Chagan, Valencia, California)
According to the manufacturer\'s instructions, slightly modified.
In particular, a cell culture of 100 μl-
Series dilution of derivative ZIKV (
10, 10 and 10)
Mix in human urine with 100 µμ l buffer AL and 12.
Qiagen enzyme solution 5 μ l.
After incubation at 56 °c for 15 min, 2.
5 μ l vector RNA solution (
Reserve concentration of 1 μg/μ l in buffer AVE)
Add 95% ml of ethanol to the lysate.
Extract RNA from each dilution in a repeating column and perform standard washing steps as recommended by the manufacturer.
RNA was eluted with 100 μ l buffer AVE at room temperature.
It is worth noting that in both extraction methods, the volume of the input and output samples is deliberately selected to be the same in order to make a fair comparison in efficiency.
RNA templates extracted from our equipment were evaluated in commercial reality
Time PCR detection system (Bio-Rad CFX-
96, Hercules, California)
Platinum One-use superscript III-
Step quantitative RT-PCR system (
TFS, Waltham, MA).
Each 20 μ l reaction consists of a mixture of 10 μ l 2 × Reaction, each 500nm for ZIKV 1086 and ZIKV 1162c primers, FAM-
Labeled hydrolysis probe (ZIKV 1107-FAM)
, And RNA templates of 2 μ l (
Primers and probe sequences see).
The specificity of ZIV primers was previously evaluated by testing dengue, West Nile, Yellow Sea, and chikungunya virus RNA.
The condition is 5 min at 50 °c (cDNA synthesis)
, 95 ° c lasts 30 seconds, 95 ° c lasts 10 seconds, and 60 ° c lasts 20 seconds [
Or known as 300 m/s s/30 m/s s/45 °f × °f (10u2009s/20u2009s)]
Real-time fluorescence detection
Time thermal cycling.
Duration of each business entitytime RT-
PCR runs for about 57 minutes.
When the speed of the commercial Hot Bike is compared to that of the hot water bottle Hot Bike, a shorter protocol is used (TTC)
Developed by the Author: 50 ℃ 30 seconds (cDNA synthesis)
, 95 ° c lasts 10 seconds, 95 ° c lasts 5 seconds, and 60 ° c lasts 10 seconds [30 u2009 s/10 u2009 s/40 u2009 × (5u2009s/10u2009s)].
The duration of the run is 36 minutes and 47 seconds. In most RT-
No PCR reaction
Template controls (NTC)
ZIKV negative controls were added in the same run.
Negative controls are templates in ZIKV negative urine samples or templates containing dengue RNA (DENV-1-4)
Extraction from dengue positive controls (
Heat-inactivated mixture of DENV-1, -2, -3 and -4 standards)
DENV from CDC-1-4 Real Time RT-PCR Assay. For the RT-
In the RPA analysis, we designed the primer and one from the real-time RT-
PCR primers and probes for detecting the prM gene of Zika virus.
RPA primers are 30 ~ according to the recommended length ~ 35 bases.
So rpa zv-F and RPA ZV-
The R primers have 31 and 30 bases respectively, and the size of the amplifier is expected to be 88 ÷ bp ().
The size of Exo stamp exo probe is 52 base lengths ()
, The first 28 bases at the end of probe 5\' overlap with rpa zv-R primer.
The 32 bases at the end of 5\' are thymine, with dT-FAM (
Fluorescent amidite)
, 33 bases at the end of 5\' are Guanyin, replaced by THF (()
, The 34 bases at the end of 5\' are thymine, which is dT-BHQ1 (
Black hole quenching Machine 1).
The design conforms to the standard that THF has at least 30 bases 5\' and THF has at least 15 bases 3.
It also meets the standard of less than 5 bases between fluorescent groups (FAM)
And quenching (BHQ1)
, There is only one secret border of the THF and BHQ1.
Spacers C3 were added at the 3\' end of the probe to prevent the extension. Real-time RT-
According to the manufacturer\'s instructions, the RPA reaction was set using the TwistAmp exo RT kit (
TwistDx, Cambridge, United Kingdom).
Optical clarity 0. 2u2009mL low-
Contour PCR strips with flat covers were used in this trial. For each 12.
5 μ l volume reaction, 420nm for each rpa zv-F and RPA ZV-
The R primer, the 120nm of the TwistAmp exo probe, and the 2 u2009 μ l of the RNA template were mixed and briefly centrifugal, and then the magnesium acetate of 14mm was added to start the reaction.
RT-in this report
RPA is first performed using fluorescence detection in the fluorescence channel (
Excitation 470 nm and detection 520 nm)
In an ESEQuant tubetta (
Konstantz, stockchagan Lake, Germany)
The procedure is maintained at 40 ° c 15 min and the measurement interval is 30 sec.
According to the instructions in the TwistDx protocol, the measurement is suspended at 4 points.
5 min, allow the removal of the reaction tube for a short stirring, then put it back into the tube scanner and immediately resume the fluorescence measurement.
Threshold time (Tt)
The first derivative of the fluorescence signal curve is used to determine the time of the constant temperature amplification reaction.
When the first derivative is greater than or equal to 0.
1. the time point is considered Tt of the analysis.
Instead of using 50 μ l in each RT-
The RPA reaction recommended by the manufacturer is only 12.
5 μ l was used to reduce the cost.
In addition, the smaller reaction volume (e. g. , 5u2009μL)
It is found that no stirring is required in order to achieve the sensitivity claimed by the manufacturer.
Although ESEQuant tubesc is a small portable device that collects fluorescent signals of up to 12 tubes over time, it is also an expensive device (
Over $7,000 per detector).
Thus, the possibility of using a mobile phone as an alternative tool for collecting fluorescent signals was explored. Real-time RT-
Execute RPA on 3D printer-
The processing ZIKV template for the main mixture was incubated using a 3D printer\'s heating bed or extruder as a heat source.
To monitor the green fluorescence of fluorescent dyes, we first tried several blue LED or UV-
The LED flashlight is used as an incentive light source, but the effect is not ideal.
So we built our own LED array (
A row of 8 led separated by 9mm)
Blue LEDs using a separate package (470u2009nm, 5-
Newark 08R2921 diameter mm. com)
As our excitation light source
This special LED model provides high strength (4.
Luminous intensity 1 cd)
Wide Perspective (30°).
The array is powered by a 9 v battery.
In order to capture the green fluorescence emitted by the blue LED lighting reaction tube, the mobile phone (Samsung Note 3)
Plastic orange filter with removal from the gel observation box (
I/O Rodeo, Inc. Pasadena, CA)
The camera placed on the camera lens takes a test tube photo every 30 seconds for $3.
95 Android camera app with lockfocus, locked-
Exposure time-
Delayed photography function (Camera FV-
5 installed on Samsung Note 3 smartphone). The time-
Visual Analysis of failed photos to identify when the signal strength of the tube is greater than ZIKV-
Urine sample control or negative NTC response signal.
We also observed the fluorescence of the tube using a blue LED Gel observation box (
Walkersville, MD).
View and record the launch using the same orange filter and camera.
After the zikv rna template is extracted by the 3D printer, the real-time RT-
PCR using a commercial thermal cycler, via RT-
PCR was performed using TTC.
TTC is a practical solution that can reducecost, simple-to-
Operation and rapid thermal cycling for underserved and developing crowd.
We showed fast PCR and RT-before-
PCR thermal cycling with TTC at a rate of 15 to 30 seconds per cycle.
TTC uses a very simple design based on \"hands-
Transfer the \"reaction tube through a series of water bath liquid to minimize the temperature rise time required for the PCR tube to reach the heat balance.
The automation of PCR reaction is controlled by the action of Arduino micro-controller and servo motor. After the RT-
After the PCR is completed, the same led and smartphone settings as described earlier can be used to view the fluorescent signals of these reaction tubes.
The construction cost per TTC is less than $200.
With TTC, many reactions were carried out in a glass capillary.
The protocol consists of a reversal of 5 sec min, followed by RT inhibition of 10 sec, followed by a degeneration cycle of 40 5 sec and an annealing/extension cycle of 30 sec, abbreviated5u2009s/30u2009s).
Recently, \"I tried a fairly good performance primer to amplify the 30 m/s/10 m/s/40 ÷ × (5u2009s/10u2009s).
It should be noted that when using a glass capillary, the main mixture prepared using the superscript III Platinum 1
Step quantitative RT-PCR system (
Life Technology)
There was no amplification produced, which may have been caused by the adsorption of reverse transcription and PCR on the wall of the glass tube, which effectively reduced the concentration of enzymes that produced the cDNA and the amplification.
So when using a glass tube in TTC, use a step-by-step PrimeScript RT-PCR Kit (
Treasure/Clontech Mountain View, CA)
Compatible with glass tube PCR reaction.
In order to provide the control of the comparison of amplification efficiency, shorter commercial agreements were also carried out.
This 30-inch s/10-inch s/40-hour x-hour (5u2009s/10u2009s)
The commercial thermal cycling reaction takes about 35 minutes to complete.
Expansion of RT-
PCR was evaluated using a 2. 2% pre-casted gel (
Flash gel for longza, volxville, MD)
7 min at 275 V (
4 μ l mixture added to 1 μ l loading dye).
The final gel image was taken with a mobile phone camera.
The typical step sizes used in the gel are 50, 100, 150, 200, 300, 500, 800, and bp.
The image j software developed by NIH quantified the fluorescence intensity of the images taken by the mobile phone camera.
The average gray scale value of the selected fluorescent region is reported.
Differences and spin between Kaijie 3D printer methods
Column method was checked.
Quantitative cycle (Cq)
Using paired t-analyze the values of the two methods
Test with online software (
QuickCalcs, La Jolla, CA for GraphPad). A -
The value is less than 0.
05 is considered to have statistical significance.
The Cq value was also analyzed by the variance equality test of Levene.
Custom message




 towell@sztuowei.com
towell@sztuowei.com